Testing hierarchical pathway kinetics with residue data on cyantraniliprole
Johannes Ranke
Last change on 6 January 2023, last compiled on 17 Februar 2023
Source:vignettes/prebuilt/2022_cyan_pathway.rmd
2022_cyan_pathway.rmdIntroduction
The purpose of this document is to test demonstrate how nonlinear hierarchical models (NLHM) based on the parent degradation models SFO, FOMC, DFOP and HS, with serial formation of two or more metabolites can be fitted with the mkin package.
It was assembled in the course of work package 1.2 of Project Number 173340 (Application of nonlinear hierarchical models to the kinetic evaluation of chemical degradation data) of the German Environment Agency carried out in 2022 and 2023.
The mkin package is used in version 1.2.3 which is currently under
development. The newly introduced functionality that is used here is a
simplification of excluding random effects for a set of fits based on a
related set of fits with a reduced model, and the documentation of the
starting parameters of the fit, so that all starting parameters of
saem fits are now listed in the summary. The
saemix package is used as a backend for fitting the NLHM,
but is also loaded to make the convergence plot function available.
This document is processed with the knitr package, which
also provides the kable function that is used to improve
the display of tabular data in R markdown documents. For parallel
processing, the parallel package is used.
library(mkin)
library(knitr)
library(saemix)
library(parallel)
n_cores <- detectCores()
if (Sys.info()["sysname"] == "Windows") {
cl <- makePSOCKcluster(n_cores)
} else {
cl <- makeForkCluster(n_cores)
}Test data
The example data are taken from the final addendum to the DAR from
2014 and are distributed with the mkin package. Residue data and time
step normalisation factors are read in using the function
read_spreadsheet from the mkin package. This function also
performs the time step normalisation.
data_file <- system.file(
"testdata", "cyantraniliprole_soil_efsa_2014.xlsx",
package = "mkin")
cyan_ds <- read_spreadsheet(data_file, parent_only = FALSE)The following tables show the covariate data and the 5 datasets that were read in from the spreadsheet file.
| pH | |
|---|---|
| Nambsheim | 7.90 |
| Tama | 6.20 |
| Gross-Umstadt | 7.04 |
| Sassafras | 4.62 |
| Lleida | 8.05 |
for (ds_name in names(cyan_ds)) {
print(
kable(mkin_long_to_wide(cyan_ds[[ds_name]]),
caption = paste("Dataset", ds_name),
booktabs = TRUE, row.names = FALSE))
cat("\n\\clearpage\n")
}| time | cyan | JCZ38 | J9C38 | JSE76 | J9Z38 |
|---|---|---|---|---|---|
| 0.000000 | 105.79 | NA | NA | NA | NA |
| 3.210424 | 77.26 | 7.92 | 11.94 | 5.58 | 9.12 |
| 7.490988 | 57.13 | 15.46 | 16.58 | 12.59 | 11.74 |
| 17.122259 | 37.74 | 15.98 | 13.36 | 26.05 | 10.77 |
| 23.543105 | 31.47 | 6.05 | 14.49 | 34.71 | 4.96 |
| 43.875788 | 16.74 | 6.07 | 7.57 | 40.38 | 6.52 |
| 67.418893 | 8.85 | 10.34 | 6.39 | 30.71 | 8.90 |
| 107.014116 | 5.19 | 9.61 | 1.95 | 20.41 | 12.93 |
| 129.487080 | 3.45 | 6.18 | 1.36 | 21.78 | 6.99 |
| 195.835832 | 2.15 | 9.13 | 0.95 | 16.29 | 7.69 |
| 254.693596 | 1.92 | 6.92 | 0.20 | 13.57 | 7.16 |
| 321.042348 | 2.26 | 7.02 | NA | 11.12 | 8.66 |
| 383.110535 | NA | 5.05 | NA | 10.64 | 5.56 |
| 0.000000 | 105.57 | NA | NA | NA | NA |
| 3.210424 | 78.88 | 12.77 | 11.94 | 5.47 | 9.12 |
| 7.490988 | 59.94 | 15.27 | 16.58 | 13.60 | 11.74 |
| 17.122259 | 39.67 | 14.26 | 13.36 | 29.44 | 10.77 |
| 23.543105 | 30.21 | 16.07 | 14.49 | 35.90 | 4.96 |
| 43.875788 | 18.06 | 9.44 | 7.57 | 42.30 | 6.52 |
| 67.418893 | 8.54 | 5.78 | 6.39 | 34.70 | 8.90 |
| 107.014116 | 7.26 | 4.54 | 1.95 | 23.33 | 12.93 |
| 129.487080 | 3.60 | 4.22 | 1.36 | 23.56 | 6.99 |
| 195.835832 | 2.84 | 3.05 | 0.95 | 16.21 | 7.69 |
| 254.693596 | 2.00 | 2.90 | 0.20 | 15.53 | 7.16 |
| 321.042348 | 1.79 | 0.94 | NA | 9.80 | 8.66 |
| 383.110535 | NA | 1.82 | NA | 9.49 | 5.56 |
| time | cyan | JCZ38 | J9Z38 | JSE76 |
|---|---|---|---|---|
| 0.000000 | 106.14 | NA | NA | NA |
| 2.400833 | 93.47 | 6.46 | 2.85 | NA |
| 5.601943 | 88.39 | 10.86 | 4.65 | 3.85 |
| 12.804442 | 72.29 | 11.97 | 4.91 | 11.24 |
| 17.606108 | 65.79 | 13.11 | 6.63 | 13.79 |
| 32.811382 | 53.16 | 11.24 | 8.90 | 23.40 |
| 50.417490 | 44.01 | 11.34 | 9.98 | 29.56 |
| 80.027761 | 33.23 | 8.82 | 11.31 | 35.63 |
| 96.833591 | 40.68 | 5.94 | 8.32 | 29.09 |
| 146.450803 | 20.65 | 4.49 | 8.72 | 36.88 |
| 190.466072 | 17.71 | 4.66 | 11.10 | 40.97 |
| 240.083284 | 14.86 | 2.27 | 11.62 | 40.11 |
| 286.499386 | 12.02 | NA | 10.73 | 42.58 |
| 0.000000 | 109.11 | NA | NA | NA |
| 2.400833 | 96.84 | 5.52 | 2.04 | 2.02 |
| 5.601943 | 85.29 | 9.65 | 2.99 | 4.39 |
| 12.804442 | 73.68 | 12.48 | 5.05 | 11.47 |
| 17.606108 | 64.89 | 12.44 | 6.29 | 15.00 |
| 32.811382 | 52.27 | 10.86 | 7.65 | 23.30 |
| 50.417490 | 42.61 | 10.54 | 9.37 | 31.06 |
| 80.027761 | 34.29 | 10.02 | 9.04 | 37.87 |
| 96.833591 | 30.50 | 6.34 | 8.14 | 33.97 |
| 146.450803 | 19.21 | 6.29 | 8.52 | 26.15 |
| 190.466072 | 17.55 | 5.81 | 9.89 | 32.08 |
| 240.083284 | 13.22 | 5.99 | 10.79 | 40.66 |
| 286.499386 | 11.09 | 6.05 | 8.82 | 42.90 |
| time | cyan | JCZ38 | J9Z38 | JSE76 |
|---|---|---|---|---|
| 0.0000000 | 103.03 | NA | NA | NA |
| 2.1014681 | 87.85 | 4.79 | 3.26 | 0.62 |
| 4.9034255 | 77.35 | 8.05 | 9.89 | 1.32 |
| 10.5073404 | 69.33 | 9.74 | 12.32 | 4.74 |
| 21.0146807 | 55.65 | 14.57 | 13.59 | 9.84 |
| 31.5220211 | 49.03 | 14.66 | 16.71 | 12.32 |
| 42.0293615 | 41.86 | 15.97 | 13.64 | 15.53 |
| 63.0440422 | 34.88 | 18.20 | 14.12 | 22.02 |
| 84.0587230 | 28.26 | 15.64 | 14.06 | 25.60 |
| 0.0000000 | 104.05 | NA | NA | NA |
| 2.1014681 | 85.25 | 2.68 | 7.32 | 0.69 |
| 4.9034255 | 77.22 | 7.28 | 8.37 | 1.45 |
| 10.5073404 | 65.23 | 10.73 | 10.93 | 4.74 |
| 21.0146807 | 57.78 | 12.29 | 14.80 | 9.05 |
| 31.5220211 | 54.83 | 14.05 | 12.01 | 11.05 |
| 42.0293615 | 45.17 | 12.12 | 17.89 | 15.71 |
| 63.0440422 | 34.83 | 12.90 | 15.86 | 22.52 |
| 84.0587230 | 26.59 | 14.28 | 14.91 | 28.48 |
| 0.0000000 | 104.62 | NA | NA | NA |
| 0.8145225 | 97.21 | NA | 4.00 | NA |
| 1.9005525 | 89.64 | 3.59 | 5.24 | NA |
| 4.0726125 | 87.90 | 4.10 | 9.58 | NA |
| 8.1452251 | 86.90 | 5.96 | 9.45 | NA |
| 12.2178376 | 74.74 | 7.83 | 15.03 | 5.33 |
| 16.2904502 | 74.13 | 8.84 | 14.41 | 5.10 |
| 24.4356753 | 65.26 | 11.84 | 18.33 | 6.71 |
| 32.5809004 | 57.70 | 12.74 | 19.93 | 9.74 |
| 0.0000000 | 101.94 | NA | NA | NA |
| 0.8145225 | 99.94 | NA | NA | NA |
| 1.9005525 | 94.87 | NA | 4.56 | NA |
| 4.0726125 | 86.96 | 6.75 | 6.90 | NA |
| 8.1452251 | 80.51 | 10.68 | 7.43 | 2.58 |
| 12.2178376 | 78.38 | 10.35 | 9.46 | 3.69 |
| 16.2904502 | 70.05 | 13.73 | 9.27 | 7.18 |
| 24.4356753 | 61.28 | 12.57 | 13.28 | 13.19 |
| 32.5809004 | 52.85 | 12.67 | 12.95 | 13.69 |
| time | cyan | JCZ38 | J9Z38 | JSE76 |
|---|---|---|---|---|
| 0.000000 | 102.17 | NA | NA | NA |
| 2.216719 | 95.49 | 1.11 | 0.10 | 0.83 |
| 5.172343 | 83.35 | 6.43 | 2.89 | 3.30 |
| 11.083593 | 78.18 | 10.00 | 5.59 | 0.81 |
| 22.167186 | 70.44 | 17.21 | 4.23 | 1.09 |
| 33.250779 | 68.00 | 20.45 | 5.86 | 1.17 |
| 44.334371 | 59.64 | 24.64 | 3.17 | 2.72 |
| 66.501557 | 50.73 | 27.50 | 6.19 | 1.27 |
| 88.668742 | 45.65 | 32.77 | 5.69 | 4.54 |
| 0.000000 | 100.43 | NA | NA | NA |
| 2.216719 | 95.34 | 3.21 | 0.14 | 0.46 |
| 5.172343 | 84.38 | 5.73 | 4.75 | 0.62 |
| 11.083593 | 78.50 | 11.89 | 3.99 | 0.73 |
| 22.167186 | 71.17 | 17.28 | 4.39 | 0.66 |
| 33.250779 | 59.41 | 18.73 | 11.85 | 2.65 |
| 44.334371 | 64.57 | 22.93 | 5.13 | 2.01 |
| 66.501557 | 49.08 | 33.39 | 5.67 | 3.63 |
| 88.668742 | 40.41 | 39.60 | 5.93 | 6.17 |
| time | cyan | JCZ38 | J9Z38 | JSE76 |
|---|---|---|---|---|
| 0.000000 | 102.71 | NA | NA | NA |
| 2.821051 | 79.11 | 5.70 | 8.07 | 0.97 |
| 6.582451 | 70.03 | 7.17 | 11.31 | 4.72 |
| 14.105253 | 50.93 | 10.25 | 14.84 | 9.95 |
| 28.210505 | 33.43 | 10.40 | 14.82 | 24.06 |
| 42.315758 | 24.69 | 9.75 | 16.38 | 29.38 |
| 56.421010 | 22.99 | 10.06 | 15.51 | 29.25 |
| 84.631516 | 14.63 | 5.63 | 14.74 | 31.04 |
| 112.842021 | 12.43 | 4.17 | 13.53 | 33.28 |
| 0.000000 | 99.31 | NA | NA | NA |
| 2.821051 | 82.07 | 6.55 | 5.60 | 1.12 |
| 6.582451 | 70.65 | 7.61 | 8.01 | 3.21 |
| 14.105253 | 53.52 | 11.48 | 10.82 | 12.24 |
| 28.210505 | 35.60 | 11.19 | 15.43 | 23.53 |
| 42.315758 | 34.26 | 11.09 | 13.26 | 27.42 |
| 56.421010 | 21.79 | 4.80 | 18.30 | 30.20 |
| 84.631516 | 14.06 | 6.30 | 16.35 | 32.32 |
| 112.842021 | 11.51 | 5.57 | 12.64 | 32.51 |
Parent only evaluations
As the pathway fits have very long run times, evaluations of the parent data are performed first, in order to determine for each hierarchical parent degradation model which random effects on the degradation model parameters are ill-defined.
cyan_sep_const <- mmkin(c("SFO", "FOMC", "DFOP", "SFORB", "HS"),
cyan_ds, quiet = TRUE, cores = n_cores)
cyan_sep_tc <- update(cyan_sep_const, error_model = "tc")
cyan_saem_full <- mhmkin(list(cyan_sep_const, cyan_sep_tc))
status(cyan_saem_full) |> kable()| const | tc | |
|---|---|---|
| SFO | OK | OK |
| FOMC | OK | OK |
| DFOP | OK | OK |
| SFORB | OK | OK |
| HS | OK | OK |
All fits converged successfully.
| const | tc | |
|---|---|---|
| SFO | sd(cyan_0) | sd(cyan_0) |
| FOMC | sd(log_beta) | sd(cyan_0) |
| DFOP | sd(cyan_0) | sd(cyan_0), sd(log_k1) |
| SFORB | sd(cyan_free_0) | sd(cyan_free_0), sd(log_k_cyan_free_bound) |
| HS | sd(cyan_0) | sd(cyan_0) |
In almost all models, the random effect for the initial concentration of the parent compound is ill-defined. For the biexponential models DFOP and SFORB, the random effect of one additional parameter is ill-defined when the two-component error model is used.
| npar | AIC | BIC | Lik | |
|---|---|---|---|---|
| SFO const | 5 | 833.9 | 832.0 | -412.0 |
| SFO tc | 6 | 831.6 | 829.3 | -409.8 |
| FOMC const | 7 | 709.1 | 706.4 | -347.6 |
| FOMC tc | 8 | 689.2 | 686.1 | -336.6 |
| DFOP const | 9 | 703.0 | 699.5 | -342.5 |
| SFORB const | 9 | 701.3 | 697.8 | -341.7 |
| HS const | 9 | 718.6 | 715.1 | -350.3 |
| DFOP tc | 10 | 703.1 | 699.2 | -341.6 |
| SFORB tc | 10 | 700.1 | 696.2 | -340.1 |
| HS tc | 10 | 716.7 | 712.8 | -348.3 |
Model comparison based on AIC and BIC indicates that the two-component error model is preferable for all parent models with the exception of DFOP. The lowest AIC and BIC values are are obtained with the FOMC model, followed by SFORB and DFOP.
Pathway fits
Evaluations with pathway established previously
To test the technical feasibility of coupling the relevant parent
degradation models with different transformation pathway models, a list
of mkinmod models is set up below. As in the EU evaluation,
parallel formation of metabolites JCZ38 and J9Z38 and secondary
formation of metabolite JSE76 from JCZ38 is used.
if (!dir.exists("cyan_dlls")) dir.create("cyan_dlls")
cyan_path_1 <- list(
sfo_path_1 = mkinmod(
cyan = mkinsub("SFO", c("JCZ38", "J9Z38")),
JCZ38 = mkinsub("SFO", "JSE76"),
J9Z38 = mkinsub("SFO"),
JSE76 = mkinsub("SFO"), quiet = TRUE,
name = "sfo_path_1", dll_dir = "cyan_dlls", overwrite = TRUE),
fomc_path_1 = mkinmod(
cyan = mkinsub("FOMC", c("JCZ38", "J9Z38")),
JCZ38 = mkinsub("SFO", "JSE76"),
J9Z38 = mkinsub("SFO"),
JSE76 = mkinsub("SFO"), quiet = TRUE,
name = "fomc_path_1", dll_dir = "cyan_dlls", overwrite = TRUE),
dfop_path_1 = mkinmod(
cyan = mkinsub("DFOP", c("JCZ38", "J9Z38")),
JCZ38 = mkinsub("SFO", "JSE76"),
J9Z38 = mkinsub("SFO"),
JSE76 = mkinsub("SFO"), quiet = TRUE,
name = "dfop_path_1", dll_dir = "cyan_dlls", overwrite = TRUE),
sforb_path_1 = mkinmod(
cyan = mkinsub("SFORB", c("JCZ38", "J9Z38")),
JCZ38 = mkinsub("SFO", "JSE76"),
J9Z38 = mkinsub("SFO"),
JSE76 = mkinsub("SFO"), quiet = TRUE,
name = "sforb_path_1", dll_dir = "cyan_dlls", overwrite = TRUE),
hs_path_1 = mkinmod(
cyan = mkinsub("HS", c("JCZ38", "J9Z38")),
JCZ38 = mkinsub("SFO", "JSE76"),
J9Z38 = mkinsub("SFO"),
JSE76 = mkinsub("SFO"), quiet = TRUE,
name = "hs_path_1", dll_dir = "cyan_dlls", overwrite = TRUE)
)To obtain suitable starting values for the NLHM fits, separate pathway fits are performed for all datasets.
f_sep_1_const <- mmkin(
cyan_path_1,
cyan_ds,
error_model = "const",
cluster = cl,
quiet = TRUE)
status(f_sep_1_const) |> kable()| Nambsheim | Tama | Gross-Umstadt | Sassafras | Lleida | |
|---|---|---|---|---|---|
| sfo_path_1 | OK | OK | OK | OK | OK |
| fomc_path_1 | OK | OK | OK | OK | OK |
| dfop_path_1 | OK | OK | OK | OK | OK |
| sforb_path_1 | OK | OK | OK | OK | OK |
| hs_path_1 | C | C | C | C | C |
| Nambsheim | Tama | Gross-Umstadt | Sassafras | Lleida | |
|---|---|---|---|---|---|
| sfo_path_1 | OK | OK | OK | OK | OK |
| fomc_path_1 | OK | OK | OK | OK | C |
| dfop_path_1 | OK | OK | OK | OK | OK |
| sforb_path_1 | OK | C | OK | OK | OK |
| hs_path_1 | C | OK | C | OK | OK |
Most separate fits converged successfully. The biggest convergence problems are seen when using the HS model with constant variance.
For the hierarchical pathway fits, those random effects that could not be quantified in the corresponding parent data analyses are excluded.
In the code below, the output of the illparms function
for the parent only fits is used as an argument
no_random_effect to the mhmkin function. The
possibility to do so was introduced in mkin version 1.2.2
which is currently under development.
f_saem_1 <- mhmkin(list(f_sep_1_const, f_sep_1_tc),
no_random_effect = illparms(cyan_saem_full),
cluster = cl)| const | tc | |
|---|---|---|
| sfo_path_1 | Fth, FO | Fth, FO |
| fomc_path_1 | OK | Fth, FO |
| dfop_path_1 | Fth, FO | Fth, FO |
| sforb_path_1 | Fth, FO | Fth, FO |
| hs_path_1 | Fth, FO | Fth, FO |
The status information from the individual fits shows that all fits
completed successfully. The matrix entries Fth and FO indicate that the
Fisher Information Matrix could not be inverted for the fixed effects
(theta) and the random effects (Omega), respectively. For the affected
fits, ill-defined parameters cannot be determined using the
illparms function, because it relies on the Fisher
Information Matrix.
| const | tc | |
|---|---|---|
| sfo_path_1 | NA | NA |
| fomc_path_1 | sd(log_k_J9Z38), sd(f_cyan_ilr_2), sd(f_JCZ38_qlogis) | NA |
| dfop_path_1 | NA | NA |
| sforb_path_1 | NA | NA |
| hs_path_1 | NA | NA |
The model comparison below suggests that the pathway fits using DFOP or SFORB for the parent compound provide the best fit.
| npar | AIC | BIC | Lik | |
|---|---|---|---|---|
| sfo_path_1 const | 16 | 2692.8 | 2686.6 | -1330.4 |
| sfo_path_1 tc | 17 | 2657.7 | 2651.1 | -1311.9 |
| fomc_path_1 const | 18 | 2427.8 | 2420.8 | -1195.9 |
| fomc_path_1 tc | 19 | 2423.4 | 2416.0 | -1192.7 |
| dfop_path_1 const | 20 | 2403.2 | 2395.4 | -1181.6 |
| sforb_path_1 const | 20 | 2401.4 | 2393.6 | -1180.7 |
| hs_path_1 const | 20 | 2427.3 | 2419.5 | -1193.7 |
| dfop_path_1 tc | 20 | 2398.0 | 2390.2 | -1179.0 |
| sforb_path_1 tc | 20 | 2399.8 | 2392.0 | -1179.9 |
| hs_path_1 tc | 21 | 2422.3 | 2414.1 | -1190.2 |
For these two parent model, successful fits are shown below. Plots of the fits with the other parent models are shown in the Appendix.
plot(f_saem_1[["dfop_path_1", "tc"]])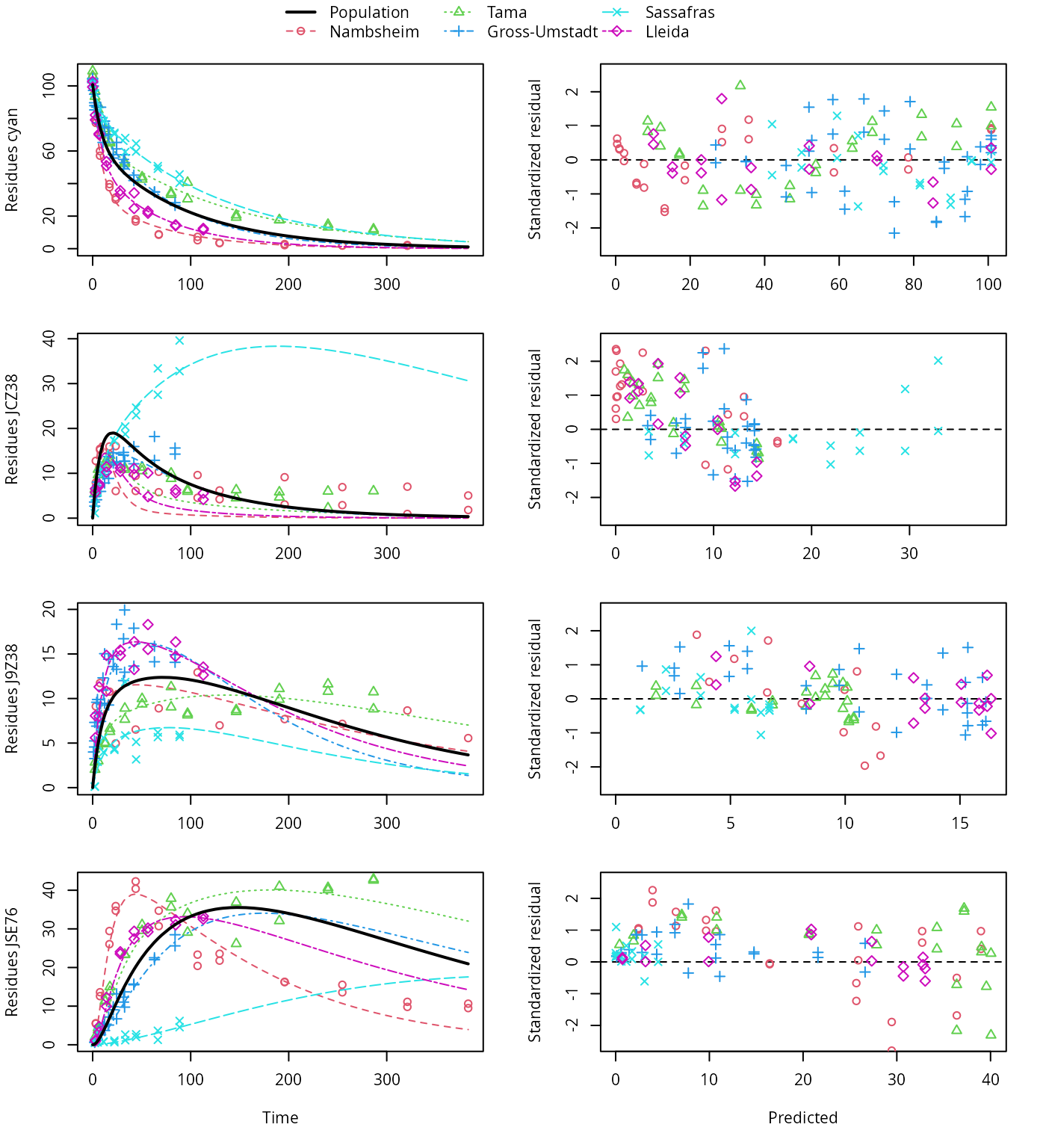
DFOP pathway fit with two-component error
plot(f_saem_1[["sforb_path_1", "tc"]])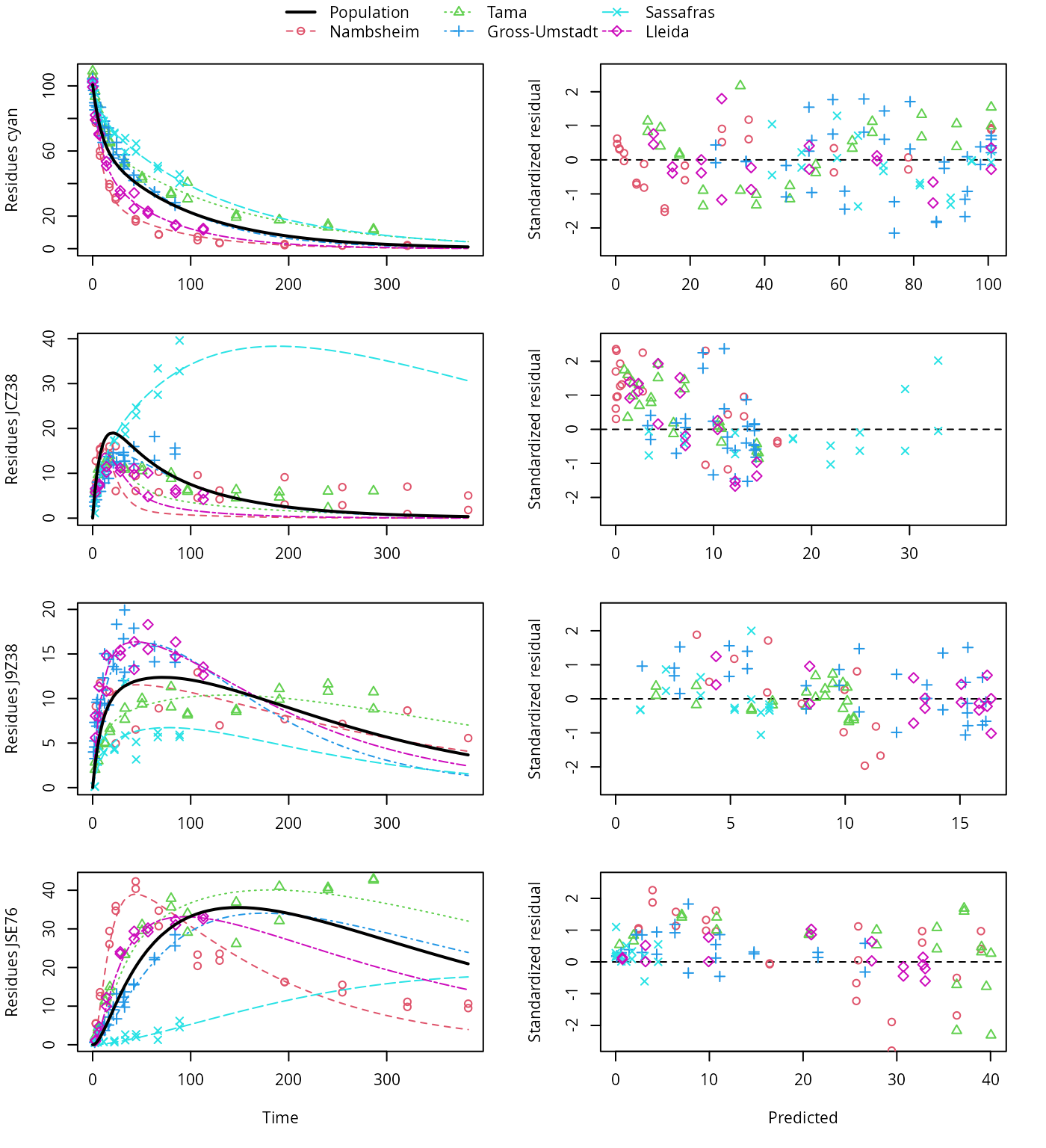
SFORB pathway fit with two-component error
A closer graphical analysis of these Figures shows that the residues of transformation product JCZ38 in the soils Tama and Nambsheim observed at later time points are strongly and systematically underestimated.
Alternative pathway fits
To improve the fit for JCZ38, a back-reaction from JSE76 to JCZ38 was introduced in an alternative version of the transformation pathway, in analogy to the back-reaction from K5A78 to K5A77. Both pairs of transformation products are pairs of an organic acid with its corresponding amide (Addendum 2014, p. 109). As FOMC provided the best fit for the parent, and the biexponential models DFOP and SFORB provided the best initial pathway fits, these three parent models are used in the alternative pathway fits.
cyan_path_2 <- list(
fomc_path_2 = mkinmod(
cyan = mkinsub("FOMC", c("JCZ38", "J9Z38")),
JCZ38 = mkinsub("SFO", "JSE76"),
J9Z38 = mkinsub("SFO"),
JSE76 = mkinsub("SFO", "JCZ38"),
name = "fomc_path_2", quiet = TRUE,
dll_dir = "cyan_dlls",
overwrite = TRUE
),
dfop_path_2 = mkinmod(
cyan = mkinsub("DFOP", c("JCZ38", "J9Z38")),
JCZ38 = mkinsub("SFO", "JSE76"),
J9Z38 = mkinsub("SFO"),
JSE76 = mkinsub("SFO", "JCZ38"),
name = "dfop_path_2", quiet = TRUE,
dll_dir = "cyan_dlls",
overwrite = TRUE
),
sforb_path_2 = mkinmod(
cyan = mkinsub("SFORB", c("JCZ38", "J9Z38")),
JCZ38 = mkinsub("SFO", "JSE76"),
J9Z38 = mkinsub("SFO"),
JSE76 = mkinsub("SFO", "JCZ38"),
name = "sforb_path_2", quiet = TRUE,
dll_dir = "cyan_dlls",
overwrite = TRUE
)
)
f_sep_2_const <- mmkin(
cyan_path_2,
cyan_ds,
error_model = "const",
cluster = cl,
quiet = TRUE)
status(f_sep_2_const) |> kable()| Nambsheim | Tama | Gross-Umstadt | Sassafras | Lleida | |
|---|---|---|---|---|---|
| fomc_path_2 | OK | OK | OK | C | OK |
| dfop_path_2 | OK | OK | OK | C | OK |
| sforb_path_2 | OK | OK | OK | C | OK |
Using constant variance, separate fits converge with the exception of the fits to the Sassafras soil data.
| Nambsheim | Tama | Gross-Umstadt | Sassafras | Lleida | |
|---|---|---|---|---|---|
| fomc_path_2 | OK | C | OK | C | OK |
| dfop_path_2 | OK | OK | OK | C | OK |
| sforb_path_2 | OK | OK | OK | OK | OK |
Using the two-component error model, all separate fits converge with the exception of the alternative pathway fit with DFOP used for the parent and the Sassafras dataset.
f_saem_2 <- mhmkin(list(f_sep_2_const, f_sep_2_tc),
no_random_effect = illparms(cyan_saem_full[2:4, ]),
cluster = cl)| const | tc | |
|---|---|---|
| fomc_path_2 | OK | FO |
| dfop_path_2 | OK | OK |
| sforb_path_2 | OK | OK |
The hierarchical fits for the alternative pathway completed successfully.
| const | tc | |
|---|---|---|
| fomc_path_2 | sd(f_JCZ38_qlogis), sd(f_JSE76_qlogis) | NA |
| dfop_path_2 | sd(f_JCZ38_qlogis), sd(f_JSE76_qlogis) | sd(f_JCZ38_qlogis), sd(f_JSE76_qlogis) |
| sforb_path_2 | sd(f_JCZ38_qlogis), sd(f_JSE76_qlogis) | sd(f_JCZ38_qlogis), sd(f_JSE76_qlogis) |
In both fits, the random effects for the formation fractions for the pathways from JCZ38 to JSE76, and for the reverse pathway from JSE76 to JCZ38 are ill-defined.
| npar | AIC | BIC | Lik | |
|---|---|---|---|---|
| fomc_path_2 const | 20 | 2308.3 | 2300.5 | -1134.2 |
| fomc_path_2 tc | 21 | 2248.3 | 2240.1 | -1103.2 |
| dfop_path_2 const | 22 | 2289.6 | 2281.0 | -1122.8 |
| sforb_path_2 const | 22 | 2284.1 | 2275.5 | -1120.0 |
| dfop_path_2 tc | 22 | 2234.4 | 2225.8 | -1095.2 |
| sforb_path_2 tc | 22 | 2240.4 | 2231.8 | -1098.2 |
The variants using the biexponential models DFOP and SFORB for the parent compound and the two-component error model give the lowest AIC and BIC values and are plotted below. Compared with the original pathway, the AIC and BIC values indicate a large improvement. This is confirmed by the plots, which show that the metabolite JCZ38 is fitted much better with this model.
plot(f_saem_2[["fomc_path_2", "tc"]])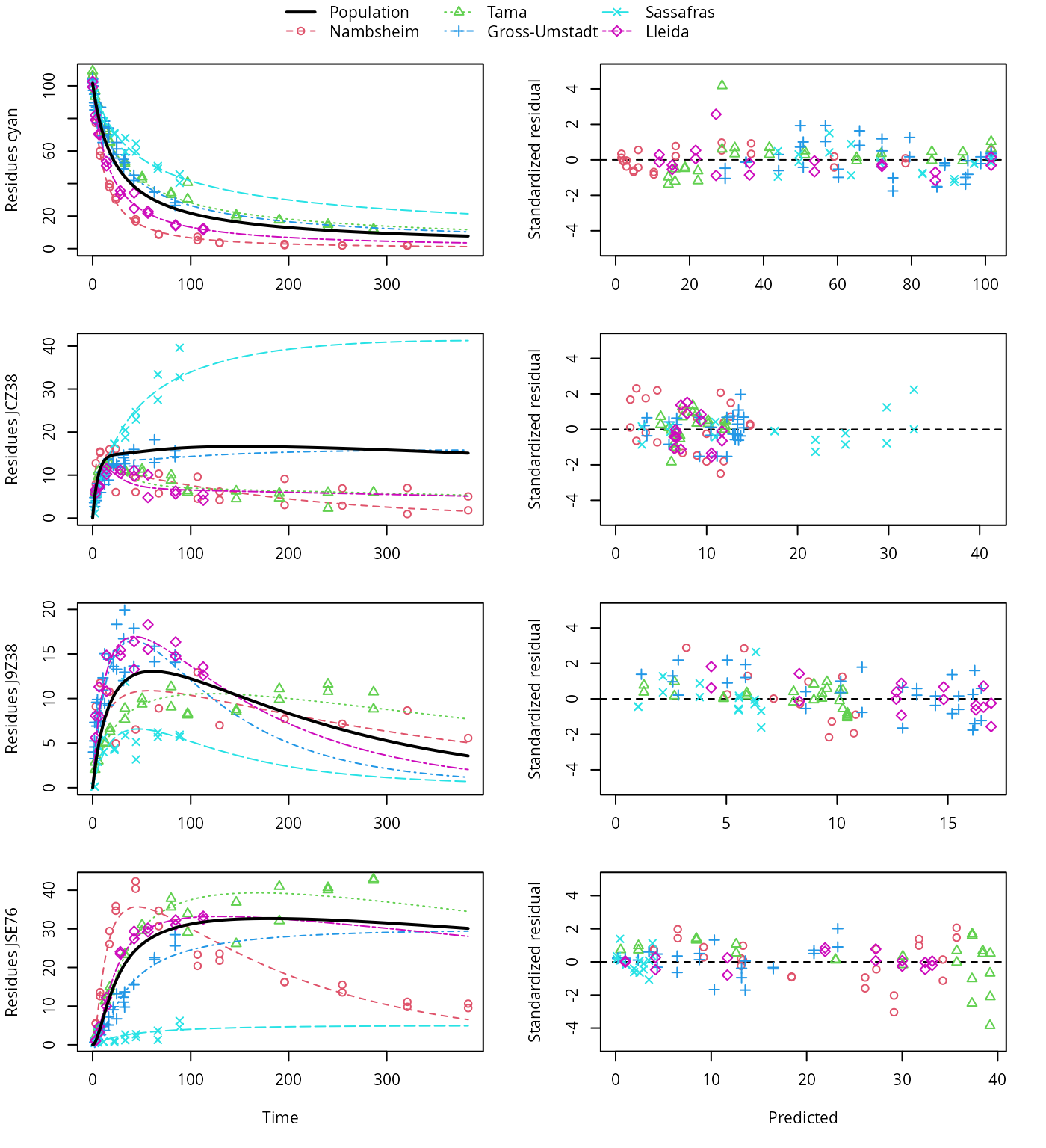
FOMC pathway fit with two-component error, alternative pathway
plot(f_saem_2[["dfop_path_2", "tc"]])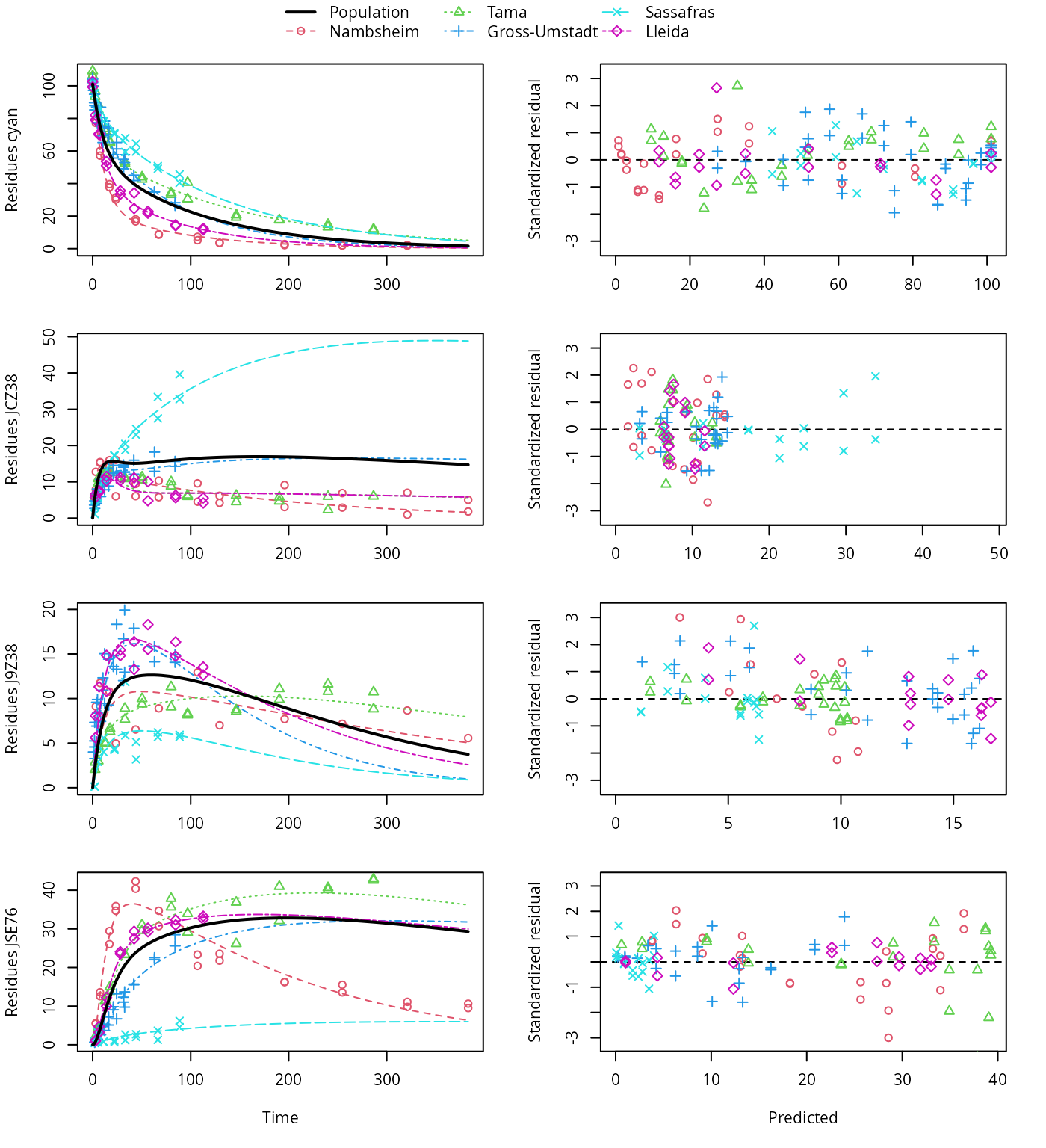
DFOP pathway fit with two-component error, alternative pathway
plot(f_saem_2[["sforb_path_2", "tc"]])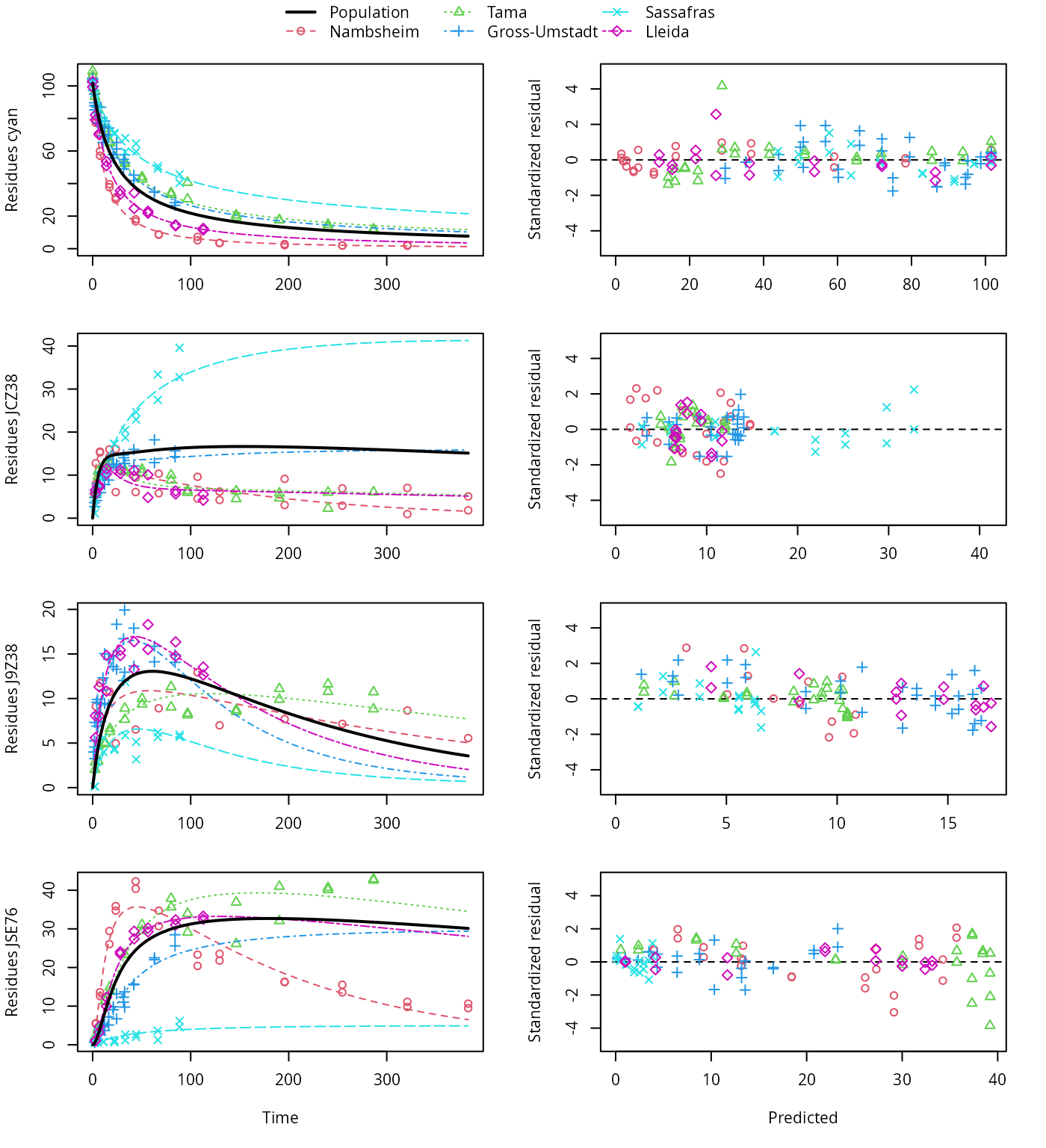
SFORB pathway fit with two-component error, alternative pathway
Refinement of alternative pathway fits
All ill-defined random effects that were identified in the parent
only fits and in the above pathway fits, are excluded for the final
evaluations below. For this purpose, a list of character vectors is
created below that can be indexed by row and column indices, and which
contains the degradation parameter names for which random effects should
be excluded for each of the hierarchical fits contained in
f_saem_2.
no_ranef <- matrix(list(), nrow = 3, ncol = 2, dimnames = dimnames(f_saem_2))
no_ranef[["fomc_path_2", "const"]] <- c("log_beta", "f_JCZ38_qlogis", "f_JSE76_qlogis")
no_ranef[["fomc_path_2", "tc"]] <- c("cyan_0", "f_JCZ38_qlogis", "f_JSE76_qlogis")
no_ranef[["dfop_path_2", "const"]] <- c("cyan_0", "f_JCZ38_qlogis", "f_JSE76_qlogis")
no_ranef[["dfop_path_2", "tc"]] <- c("cyan_0", "log_k1", "f_JCZ38_qlogis", "f_JSE76_qlogis")
no_ranef[["sforb_path_2", "const"]] <- c("cyan_free_0",
"f_JCZ38_qlogis", "f_JSE76_qlogis")
no_ranef[["sforb_path_2", "tc"]] <- c("cyan_free_0", "log_k_cyan_free_bound",
"f_JCZ38_qlogis", "f_JSE76_qlogis")
clusterExport(cl, "no_ranef")
f_saem_3 <- update(f_saem_2,
no_random_effect = no_ranef,
cluster = cl)| const | tc | |
|---|---|---|
| fomc_path_2 | E | Fth |
| dfop_path_2 | Fth | Fth |
| sforb_path_2 | Fth | Fth |
With the exception of the FOMC pathway fit with constant variance, all updated fits completed successfully. However, the Fisher Information Matrix for the fixed effects (Fth) could not be inverted, so no confidence intervals for the optimised parameters are available.
| const | tc | |
|---|---|---|
| fomc_path_2 | E | |
| dfop_path_2 | ||
| sforb_path_2 |
| npar | AIC | BIC | Lik | |
|---|---|---|---|---|
| fomc_path_2 tc | 19 | 2250.9 | 2243.5 | -1106.5 |
| dfop_path_2 const | 20 | 2281.7 | 2273.9 | -1120.8 |
| sforb_path_2 const | 20 | 2279.5 | 2271.7 | -1119.7 |
| dfop_path_2 tc | 20 | 2231.5 | 2223.7 | -1095.8 |
| sforb_path_2 tc | 20 | 2235.7 | 2227.9 | -1097.9 |
While the AIC and BIC values of the best fit (DFOP pathway fit with two-component error) are lower than in the previous fits with the alternative pathway, the practical value of these refined evaluations is limited as no confidence intervals are obtained.
Conclusion
It was demonstrated that a relatively complex transformation pathway with parallel formation of two primary metabolites and one secondary metabolite can be fitted even if the data in the individual datasets are quite different and partly only cover the formation phase.
The run times of the pathway fits were several hours, limiting the practical feasibility of iterative refinements based on ill-defined parameters and of alternative checks of parameter identifiability based on multistart runs.
Acknowledgements
The helpful comments by Janina Wöltjen of the German Environment Agency are gratefully acknowledged.
Appendix
Plots of fits that were not refined further
plot(f_saem_1[["sfo_path_1", "tc"]])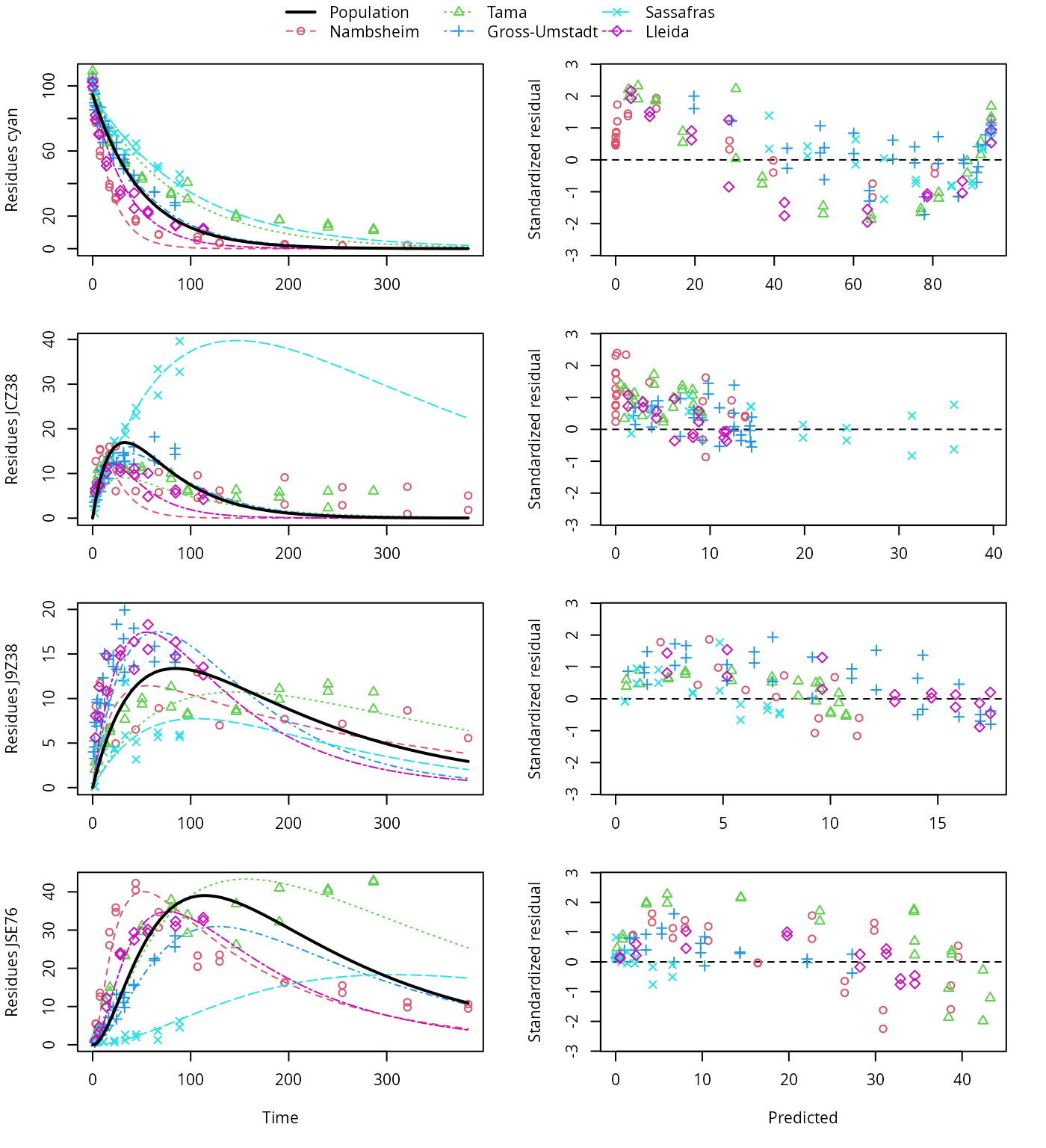
SFO pathway fit with two-component error
plot(f_saem_1[["fomc_path_1", "tc"]])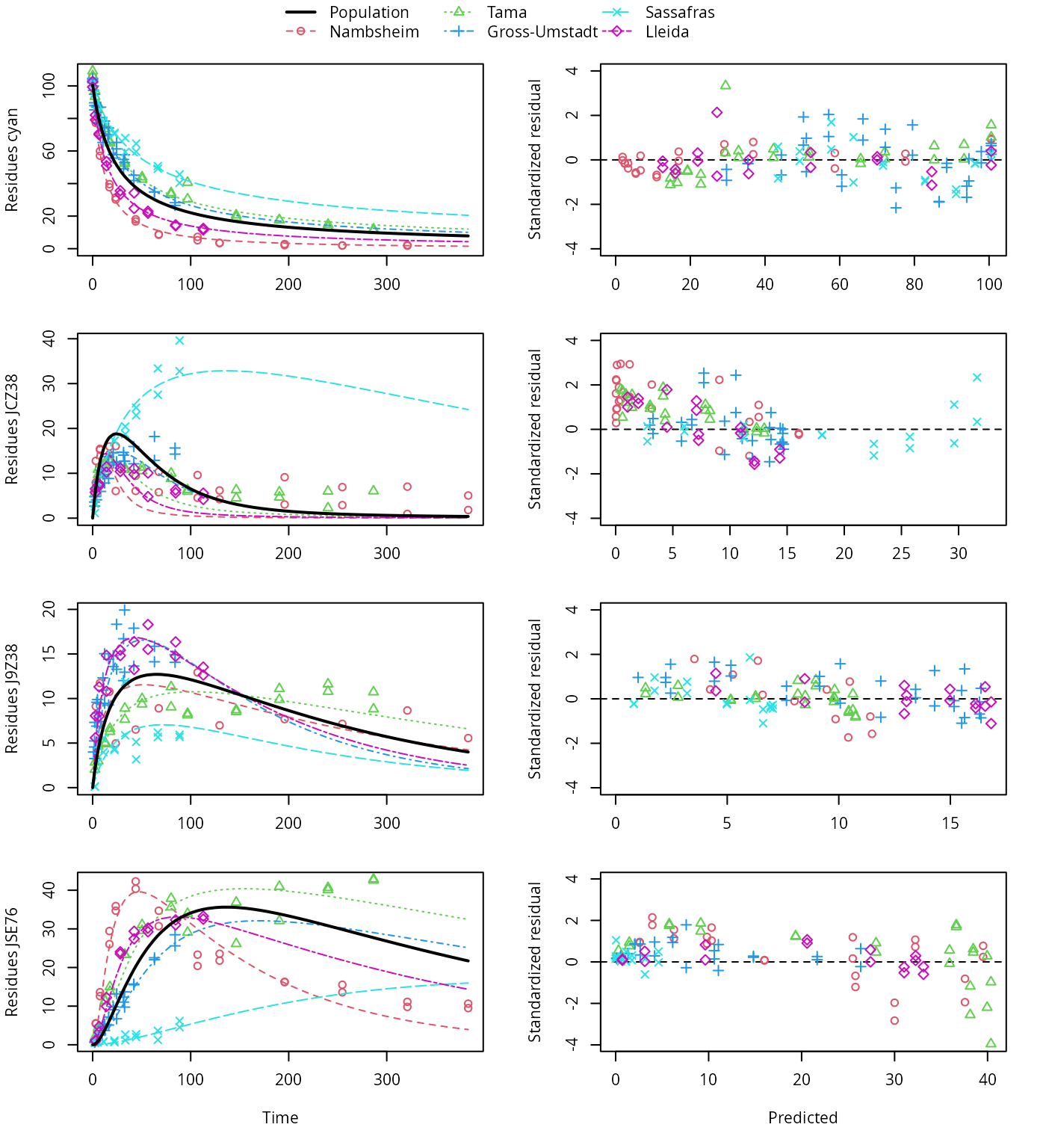
FOMC pathway fit with two-component error
plot(f_saem_1[["sforb_path_1", "tc"]])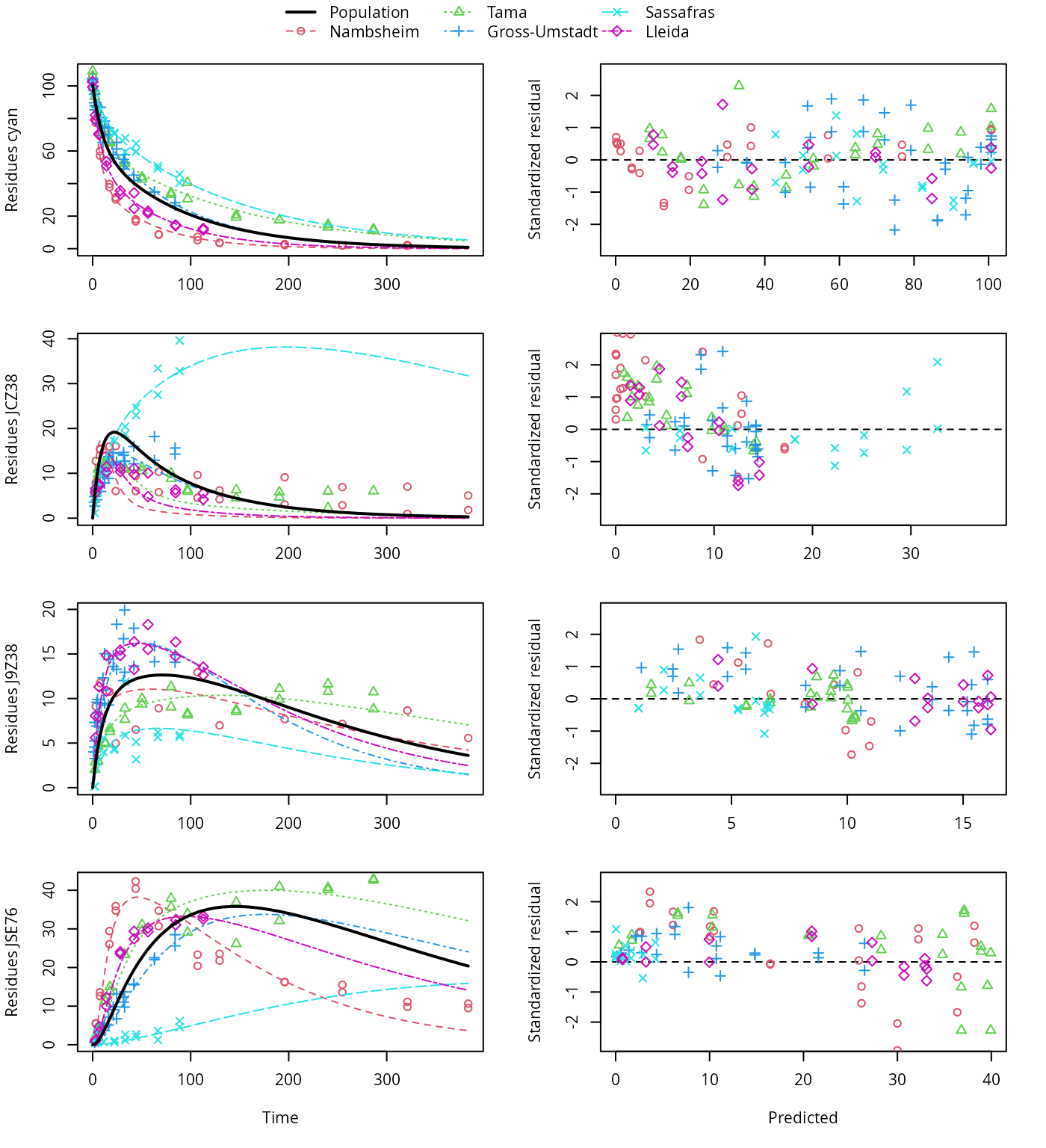
HS pathway fit with two-component error
Hierarchical fit listings
Pathway 1
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:07:38 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - k_cyan * cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * k_cyan * cyan - k_JCZ38 * JCZ38
d_J9Z38/dt = + f_cyan_to_J9Z38 * k_cyan * cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1088.473 s
Using 300, 100 iterations and 10 chains
Variance model: Constant variance
Starting values for degradation parameters:
cyan_0 log_k_cyan log_k_JCZ38 log_k_J9Z38 log_k_JSE76
95.3304 -3.8459 -3.1305 -5.0678 -5.3196
f_cyan_ilr_1 f_cyan_ilr_2 f_JCZ38_qlogis
0.8158 22.5404 10.4289
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_cyan log_k_JCZ38 log_k_J9Z38 log_k_JSE76
cyan_0 4.797 0.0000 0.000 0.000 0.0000
log_k_cyan 0.000 0.9619 0.000 0.000 0.0000
log_k_JCZ38 0.000 0.0000 2.139 0.000 0.0000
log_k_J9Z38 0.000 0.0000 0.000 1.639 0.0000
log_k_JSE76 0.000 0.0000 0.000 0.000 0.7894
f_cyan_ilr_1 0.000 0.0000 0.000 0.000 0.0000
f_cyan_ilr_2 0.000 0.0000 0.000 0.000 0.0000
f_JCZ38_qlogis 0.000 0.0000 0.000 0.000 0.0000
f_cyan_ilr_1 f_cyan_ilr_2 f_JCZ38_qlogis
cyan_0 0.0000 0.000 0.00
log_k_cyan 0.0000 0.000 0.00
log_k_JCZ38 0.0000 0.000 0.00
log_k_J9Z38 0.0000 0.000 0.00
log_k_JSE76 0.0000 0.000 0.00
f_cyan_ilr_1 0.7714 0.000 0.00
f_cyan_ilr_2 0.0000 8.684 0.00
f_JCZ38_qlogis 0.0000 0.000 13.48
Starting values for error model parameters:
a.1
1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2693 2687 -1330
Optimised parameters:
est. lower upper
cyan_0 95.0946 NA NA
log_k_cyan -3.8544 NA NA
log_k_JCZ38 -3.0402 NA NA
log_k_J9Z38 -5.0109 NA NA
log_k_JSE76 -5.2857 NA NA
f_cyan_ilr_1 0.8069 NA NA
f_cyan_ilr_2 16.6623 NA NA
f_JCZ38_qlogis 1.3602 NA NA
a.1 4.8326 NA NA
SD.log_k_cyan 0.5842 NA NA
SD.log_k_JCZ38 1.2680 NA NA
SD.log_k_J9Z38 0.3626 NA NA
SD.log_k_JSE76 0.5244 NA NA
SD.f_cyan_ilr_1 0.2752 NA NA
SD.f_cyan_ilr_2 2.3556 NA NA
SD.f_JCZ38_qlogis 0.2400 NA NA
Correlation is not available
Random effects:
est. lower upper
SD.log_k_cyan 0.5842 NA NA
SD.log_k_JCZ38 1.2680 NA NA
SD.log_k_J9Z38 0.3626 NA NA
SD.log_k_JSE76 0.5244 NA NA
SD.f_cyan_ilr_1 0.2752 NA NA
SD.f_cyan_ilr_2 2.3556 NA NA
SD.f_JCZ38_qlogis 0.2400 NA NA
Variance model:
est. lower upper
a.1 4.833 NA NA
Backtransformed parameters:
est. lower upper
cyan_0 95.094581 NA NA
k_cyan 0.021186 NA NA
k_JCZ38 0.047825 NA NA
k_J9Z38 0.006665 NA NA
k_JSE76 0.005063 NA NA
f_cyan_to_JCZ38 0.757885 NA NA
f_cyan_to_J9Z38 0.242115 NA NA
f_JCZ38_to_JSE76 0.795792 NA NA
Resulting formation fractions:
ff
cyan_JCZ38 7.579e-01
cyan_J9Z38 2.421e-01
cyan_sink 5.877e-10
JCZ38_JSE76 7.958e-01
JCZ38_sink 2.042e-01
Estimated disappearance times:
DT50 DT90
cyan 32.72 108.68
JCZ38 14.49 48.15
J9Z38 103.99 345.46
JSE76 136.90 454.76
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:08:17 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - k_cyan * cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * k_cyan * cyan - k_JCZ38 * JCZ38
d_J9Z38/dt = + f_cyan_to_J9Z38 * k_cyan * cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1127.552 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_0 log_k_cyan log_k_JCZ38 log_k_J9Z38 log_k_JSE76
96.0039 -3.8907 -3.1276 -5.0069 -4.9367
f_cyan_ilr_1 f_cyan_ilr_2 f_JCZ38_qlogis
0.7937 20.0030 15.1336
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_cyan log_k_JCZ38 log_k_J9Z38 log_k_JSE76
cyan_0 4.859 0.000 0.00 0.00 0.0000
log_k_cyan 0.000 0.962 0.00 0.00 0.0000
log_k_JCZ38 0.000 0.000 2.04 0.00 0.0000
log_k_J9Z38 0.000 0.000 0.00 1.72 0.0000
log_k_JSE76 0.000 0.000 0.00 0.00 0.9076
f_cyan_ilr_1 0.000 0.000 0.00 0.00 0.0000
f_cyan_ilr_2 0.000 0.000 0.00 0.00 0.0000
f_JCZ38_qlogis 0.000 0.000 0.00 0.00 0.0000
f_cyan_ilr_1 f_cyan_ilr_2 f_JCZ38_qlogis
cyan_0 0.0000 0.000 0.00
log_k_cyan 0.0000 0.000 0.00
log_k_JCZ38 0.0000 0.000 0.00
log_k_J9Z38 0.0000 0.000 0.00
log_k_JSE76 0.0000 0.000 0.00
f_cyan_ilr_1 0.7598 0.000 0.00
f_cyan_ilr_2 0.0000 7.334 0.00
f_JCZ38_qlogis 0.0000 0.000 11.78
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2658 2651 -1312
Optimised parameters:
est. lower upper
cyan_0 94.72923 NA NA
log_k_cyan -3.91670 NA NA
log_k_JCZ38 -3.12917 NA NA
log_k_J9Z38 -5.06070 NA NA
log_k_JSE76 -5.09254 NA NA
f_cyan_ilr_1 0.81116 NA NA
f_cyan_ilr_2 39.97850 NA NA
f_JCZ38_qlogis 3.09728 NA NA
a.1 3.95044 NA NA
b.1 0.07998 NA NA
SD.log_k_cyan 0.58855 NA NA
SD.log_k_JCZ38 1.29753 NA NA
SD.log_k_J9Z38 0.62851 NA NA
SD.log_k_JSE76 0.37235 NA NA
SD.f_cyan_ilr_1 0.37346 NA NA
SD.f_cyan_ilr_2 1.41667 NA NA
SD.f_JCZ38_qlogis 1.81467 NA NA
Correlation is not available
Random effects:
est. lower upper
SD.log_k_cyan 0.5886 NA NA
SD.log_k_JCZ38 1.2975 NA NA
SD.log_k_J9Z38 0.6285 NA NA
SD.log_k_JSE76 0.3724 NA NA
SD.f_cyan_ilr_1 0.3735 NA NA
SD.f_cyan_ilr_2 1.4167 NA NA
SD.f_JCZ38_qlogis 1.8147 NA NA
Variance model:
est. lower upper
a.1 3.95044 NA NA
b.1 0.07998 NA NA
Backtransformed parameters:
est. lower upper
cyan_0 94.729229 NA NA
k_cyan 0.019907 NA NA
k_JCZ38 0.043754 NA NA
k_J9Z38 0.006341 NA NA
k_JSE76 0.006142 NA NA
f_cyan_to_JCZ38 0.758991 NA NA
f_cyan_to_J9Z38 0.241009 NA NA
f_JCZ38_to_JSE76 0.956781 NA NA
Resulting formation fractions:
ff
cyan_JCZ38 0.75899
cyan_J9Z38 0.24101
cyan_sink 0.00000
JCZ38_JSE76 0.95678
JCZ38_sink 0.04322
Estimated disappearance times:
DT50 DT90
cyan 34.82 115.67
JCZ38 15.84 52.63
J9Z38 109.31 363.12
JSE76 112.85 374.87
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:09:12 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - (alpha/beta) * 1/((time/beta) + 1) * cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * (alpha/beta) * 1/((time/beta) + 1) *
cyan - k_JCZ38 * JCZ38
d_J9Z38/dt = + f_cyan_to_J9Z38 * (alpha/beta) * 1/((time/beta) + 1) *
cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1182.258 s
Using 300, 100 iterations and 10 chains
Variance model: Constant variance
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
101.2314 -3.3680 -5.1108 -5.9416 0.7144
f_cyan_ilr_2 f_JCZ38_qlogis log_alpha log_beta
7.3870 15.7604 -0.1791 2.9811
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 5.416 0.000 0.0 0.000 0.0000
log_k_JCZ38 0.000 2.439 0.0 0.000 0.0000
log_k_J9Z38 0.000 0.000 1.7 0.000 0.0000
log_k_JSE76 0.000 0.000 0.0 1.856 0.0000
f_cyan_ilr_1 0.000 0.000 0.0 0.000 0.7164
f_cyan_ilr_2 0.000 0.000 0.0 0.000 0.0000
f_JCZ38_qlogis 0.000 0.000 0.0 0.000 0.0000
log_alpha 0.000 0.000 0.0 0.000 0.0000
log_beta 0.000 0.000 0.0 0.000 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis log_alpha log_beta
cyan_0 0.00 0.00 0.0000 0.0000
log_k_JCZ38 0.00 0.00 0.0000 0.0000
log_k_J9Z38 0.00 0.00 0.0000 0.0000
log_k_JSE76 0.00 0.00 0.0000 0.0000
f_cyan_ilr_1 0.00 0.00 0.0000 0.0000
f_cyan_ilr_2 12.33 0.00 0.0000 0.0000
f_JCZ38_qlogis 0.00 20.42 0.0000 0.0000
log_alpha 0.00 0.00 0.4144 0.0000
log_beta 0.00 0.00 0.0000 0.5077
Starting values for error model parameters:
a.1
1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2428 2421 -1196
Optimised parameters:
est. lower upper
cyan_0 101.0225 98.306270 103.7387
log_k_JCZ38 -3.3786 -4.770657 -1.9866
log_k_J9Z38 -5.2603 -5.902085 -4.6186
log_k_JSE76 -6.1427 -7.318336 -4.9671
f_cyan_ilr_1 0.7437 0.421215 1.0663
f_cyan_ilr_2 0.9108 0.267977 1.5537
f_JCZ38_qlogis 2.0487 0.524897 3.5724
log_alpha -0.2268 -0.618049 0.1644
log_beta 2.8986 2.700701 3.0964
a.1 3.4058 3.169913 3.6416
SD.cyan_0 2.5279 0.454190 4.6016
SD.log_k_JCZ38 1.5636 0.572824 2.5543
SD.log_k_J9Z38 0.5316 -0.004405 1.0677
SD.log_k_JSE76 0.9903 0.106325 1.8742
SD.f_cyan_ilr_1 0.3464 0.112066 0.5807
SD.f_cyan_ilr_2 0.2804 -0.393900 0.9546
SD.f_JCZ38_qlogis 0.9416 -0.152986 2.0362
SD.log_alpha 0.4273 0.161044 0.6936
Correlation:
cyan_0 l__JCZ3 l__J9Z3 l__JSE7 f_cy__1 f_cy__2 f_JCZ38 log_lph
log_k_JCZ38 -0.0156
log_k_J9Z38 -0.0493 0.0073
log_k_JSE76 -0.0329 0.0018 0.0069
f_cyan_ilr_1 -0.0086 0.0180 -0.1406 0.0012
f_cyan_ilr_2 -0.2629 0.0779 0.2826 0.0274 0.0099
f_JCZ38_qlogis 0.0713 -0.0747 -0.0505 0.1169 -0.1022 -0.4893
log_alpha -0.0556 0.0120 0.0336 0.0193 0.0036 0.0840 -0.0489
log_beta -0.2898 0.0460 0.1305 0.0768 0.0190 0.4071 -0.1981 0.2772
Random effects:
est. lower upper
SD.cyan_0 2.5279 0.454190 4.6016
SD.log_k_JCZ38 1.5636 0.572824 2.5543
SD.log_k_J9Z38 0.5316 -0.004405 1.0677
SD.log_k_JSE76 0.9903 0.106325 1.8742
SD.f_cyan_ilr_1 0.3464 0.112066 0.5807
SD.f_cyan_ilr_2 0.2804 -0.393900 0.9546
SD.f_JCZ38_qlogis 0.9416 -0.152986 2.0362
SD.log_alpha 0.4273 0.161044 0.6936
Variance model:
est. lower upper
a.1 3.406 3.17 3.642
Backtransformed parameters:
est. lower upper
cyan_0 1.010e+02 9.831e+01 1.037e+02
k_JCZ38 3.409e-02 8.475e-03 1.372e-01
k_J9Z38 5.194e-03 2.734e-03 9.867e-03
k_JSE76 2.149e-03 6.633e-04 6.963e-03
f_cyan_to_JCZ38 6.481e-01 NA NA
f_cyan_to_J9Z38 2.264e-01 NA NA
f_JCZ38_to_JSE76 8.858e-01 6.283e-01 9.727e-01
alpha 7.971e-01 5.390e-01 1.179e+00
beta 1.815e+01 1.489e+01 2.212e+01
Resulting formation fractions:
ff
cyan_JCZ38 0.6481
cyan_J9Z38 0.2264
cyan_sink 0.1255
JCZ38_JSE76 0.8858
JCZ38_sink 0.1142
Estimated disappearance times:
DT50 DT90 DT50back
cyan 25.15 308.01 92.72
JCZ38 20.33 67.54 NA
J9Z38 133.46 443.35 NA
JSE76 322.53 1071.42 NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:09:18 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - (alpha/beta) * 1/((time/beta) + 1) * cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * (alpha/beta) * 1/((time/beta) + 1) *
cyan - k_JCZ38 * JCZ38
d_J9Z38/dt = + f_cyan_to_J9Z38 * (alpha/beta) * 1/((time/beta) + 1) *
cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1188.041 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
101.13827 -3.32493 -5.08921 -5.93478 0.71330
f_cyan_ilr_2 f_JCZ38_qlogis log_alpha log_beta
10.05989 12.79248 -0.09621 3.10646
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 5.643 0.000 0.000 0.00 0.0000
log_k_JCZ38 0.000 2.319 0.000 0.00 0.0000
log_k_J9Z38 0.000 0.000 1.731 0.00 0.0000
log_k_JSE76 0.000 0.000 0.000 1.86 0.0000
f_cyan_ilr_1 0.000 0.000 0.000 0.00 0.7186
f_cyan_ilr_2 0.000 0.000 0.000 0.00 0.0000
f_JCZ38_qlogis 0.000 0.000 0.000 0.00 0.0000
log_alpha 0.000 0.000 0.000 0.00 0.0000
log_beta 0.000 0.000 0.000 0.00 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis log_alpha log_beta
cyan_0 0.00 0.00 0.0000 0.0000
log_k_JCZ38 0.00 0.00 0.0000 0.0000
log_k_J9Z38 0.00 0.00 0.0000 0.0000
log_k_JSE76 0.00 0.00 0.0000 0.0000
f_cyan_ilr_1 0.00 0.00 0.0000 0.0000
f_cyan_ilr_2 12.49 0.00 0.0000 0.0000
f_JCZ38_qlogis 0.00 20.19 0.0000 0.0000
log_alpha 0.00 0.00 0.3142 0.0000
log_beta 0.00 0.00 0.0000 0.7331
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2423 2416 -1193
Optimised parameters:
est. lower upper
cyan_0 100.57649 NA NA
log_k_JCZ38 -3.46250 NA NA
log_k_J9Z38 -5.24442 NA NA
log_k_JSE76 -5.75229 NA NA
f_cyan_ilr_1 0.68480 NA NA
f_cyan_ilr_2 0.61670 NA NA
f_JCZ38_qlogis 87.97407 NA NA
log_alpha -0.15699 NA NA
log_beta 3.01540 NA NA
a.1 3.11518 NA NA
b.1 0.04445 NA NA
SD.log_k_JCZ38 1.40732 NA NA
SD.log_k_J9Z38 0.56510 NA NA
SD.log_k_JSE76 0.72067 NA NA
SD.f_cyan_ilr_1 0.31199 NA NA
SD.f_cyan_ilr_2 0.36894 NA NA
SD.f_JCZ38_qlogis 6.92892 NA NA
SD.log_alpha 0.25662 NA NA
SD.log_beta 0.35845 NA NA
Correlation is not available
Random effects:
est. lower upper
SD.log_k_JCZ38 1.4073 NA NA
SD.log_k_J9Z38 0.5651 NA NA
SD.log_k_JSE76 0.7207 NA NA
SD.f_cyan_ilr_1 0.3120 NA NA
SD.f_cyan_ilr_2 0.3689 NA NA
SD.f_JCZ38_qlogis 6.9289 NA NA
SD.log_alpha 0.2566 NA NA
SD.log_beta 0.3585 NA NA
Variance model:
est. lower upper
a.1 3.11518 NA NA
b.1 0.04445 NA NA
Backtransformed parameters:
est. lower upper
cyan_0 1.006e+02 NA NA
k_JCZ38 3.135e-02 NA NA
k_J9Z38 5.277e-03 NA NA
k_JSE76 3.175e-03 NA NA
f_cyan_to_JCZ38 5.991e-01 NA NA
f_cyan_to_J9Z38 2.275e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
alpha 8.547e-01 NA NA
beta 2.040e+01 NA NA
Resulting formation fractions:
ff
cyan_JCZ38 0.5991
cyan_J9Z38 0.2275
cyan_sink 0.1734
JCZ38_JSE76 1.0000
JCZ38_sink 0.0000
Estimated disappearance times:
DT50 DT90 DT50back
cyan 25.50 281.29 84.68
JCZ38 22.11 73.44 NA
J9Z38 131.36 436.35 NA
JSE76 218.28 725.11 NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:10:30 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 *
time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time)))
* cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_JCZ38 * JCZ38
d_J9Z38/dt = + f_cyan_to_J9Z38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1260.905 s
Using 300, 100 iterations and 10 chains
Variance model: Constant variance
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
102.0644 -3.4008 -5.0024 -5.8613 0.6855
f_cyan_ilr_2 f_JCZ38_qlogis log_k1 log_k2 g_qlogis
1.2365 13.7245 -1.8641 -4.5063 -0.6468
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 4.466 0.000 0.000 0.000 0.0000
log_k_JCZ38 0.000 2.382 0.000 0.000 0.0000
log_k_J9Z38 0.000 0.000 1.595 0.000 0.0000
log_k_JSE76 0.000 0.000 0.000 1.245 0.0000
f_cyan_ilr_1 0.000 0.000 0.000 0.000 0.6852
f_cyan_ilr_2 0.000 0.000 0.000 0.000 0.0000
f_JCZ38_qlogis 0.000 0.000 0.000 0.000 0.0000
log_k1 0.000 0.000 0.000 0.000 0.0000
log_k2 0.000 0.000 0.000 0.000 0.0000
g_qlogis 0.000 0.000 0.000 0.000 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis log_k1 log_k2 g_qlogis
cyan_0 0.00 0.00 0.0000 0.0000 0.000
log_k_JCZ38 0.00 0.00 0.0000 0.0000 0.000
log_k_J9Z38 0.00 0.00 0.0000 0.0000 0.000
log_k_JSE76 0.00 0.00 0.0000 0.0000 0.000
f_cyan_ilr_1 0.00 0.00 0.0000 0.0000 0.000
f_cyan_ilr_2 1.28 0.00 0.0000 0.0000 0.000
f_JCZ38_qlogis 0.00 16.11 0.0000 0.0000 0.000
log_k1 0.00 0.00 0.9866 0.0000 0.000
log_k2 0.00 0.00 0.0000 0.5953 0.000
g_qlogis 0.00 0.00 0.0000 0.0000 1.583
Starting values for error model parameters:
a.1
1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2403 2395 -1182
Optimised parameters:
est. lower upper
cyan_0 102.6079 NA NA
log_k_JCZ38 -3.4855 NA NA
log_k_J9Z38 -5.1686 NA NA
log_k_JSE76 -5.6697 NA NA
f_cyan_ilr_1 0.6714 NA NA
f_cyan_ilr_2 0.4986 NA NA
f_JCZ38_qlogis 55.4760 NA NA
log_k1 -1.8409 NA NA
log_k2 -4.4915 NA NA
g_qlogis -0.6403 NA NA
a.1 3.2387 NA NA
SD.log_k_JCZ38 1.4524 NA NA
SD.log_k_J9Z38 0.5151 NA NA
SD.log_k_JSE76 0.6514 NA NA
SD.f_cyan_ilr_1 0.3023 NA NA
SD.f_cyan_ilr_2 0.2959 NA NA
SD.f_JCZ38_qlogis 1.9984 NA NA
SD.log_k1 0.5188 NA NA
SD.log_k2 0.3894 NA NA
SD.g_qlogis 0.8579 NA NA
Correlation is not available
Random effects:
est. lower upper
SD.log_k_JCZ38 1.4524 NA NA
SD.log_k_J9Z38 0.5151 NA NA
SD.log_k_JSE76 0.6514 NA NA
SD.f_cyan_ilr_1 0.3023 NA NA
SD.f_cyan_ilr_2 0.2959 NA NA
SD.f_JCZ38_qlogis 1.9984 NA NA
SD.log_k1 0.5188 NA NA
SD.log_k2 0.3894 NA NA
SD.g_qlogis 0.8579 NA NA
Variance model:
est. lower upper
a.1 3.239 NA NA
Backtransformed parameters:
est. lower upper
cyan_0 1.026e+02 NA NA
k_JCZ38 3.064e-02 NA NA
k_J9Z38 5.692e-03 NA NA
k_JSE76 3.449e-03 NA NA
f_cyan_to_JCZ38 5.798e-01 NA NA
f_cyan_to_J9Z38 2.243e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
k1 1.587e-01 NA NA
k2 1.120e-02 NA NA
g 3.452e-01 NA NA
Resulting formation fractions:
ff
cyan_JCZ38 0.5798
cyan_J9Z38 0.2243
cyan_sink 0.1958
JCZ38_JSE76 1.0000
JCZ38_sink 0.0000
Estimated disappearance times:
DT50 DT90 DT50back DT50_k1 DT50_k2
cyan 25.21 167.73 50.49 4.368 61.87
JCZ38 22.62 75.15 NA NA NA
J9Z38 121.77 404.50 NA NA NA
JSE76 200.98 667.64 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:16:28 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 *
time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time)))
* cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_JCZ38 * JCZ38
d_J9Z38/dt = + f_cyan_to_J9Z38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1617.774 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
101.3964 -3.3626 -4.9792 -5.8727 0.6814
f_cyan_ilr_2 f_JCZ38_qlogis log_k1 log_k2 g_qlogis
6.7799 13.7245 -1.9222 -4.5035 -0.7172
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 5.317 0.000 0.000 0.000 0.0000
log_k_JCZ38 0.000 2.272 0.000 0.000 0.0000
log_k_J9Z38 0.000 0.000 1.633 0.000 0.0000
log_k_JSE76 0.000 0.000 0.000 1.271 0.0000
f_cyan_ilr_1 0.000 0.000 0.000 0.000 0.6838
f_cyan_ilr_2 0.000 0.000 0.000 0.000 0.0000
f_JCZ38_qlogis 0.000 0.000 0.000 0.000 0.0000
log_k1 0.000 0.000 0.000 0.000 0.0000
log_k2 0.000 0.000 0.000 0.000 0.0000
g_qlogis 0.000 0.000 0.000 0.000 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis log_k1 log_k2 g_qlogis
cyan_0 0.00 0.00 0.0000 0.0000 0.000
log_k_JCZ38 0.00 0.00 0.0000 0.0000 0.000
log_k_J9Z38 0.00 0.00 0.0000 0.0000 0.000
log_k_JSE76 0.00 0.00 0.0000 0.0000 0.000
f_cyan_ilr_1 0.00 0.00 0.0000 0.0000 0.000
f_cyan_ilr_2 11.77 0.00 0.0000 0.0000 0.000
f_JCZ38_qlogis 0.00 16.11 0.0000 0.0000 0.000
log_k1 0.00 0.00 0.9496 0.0000 0.000
log_k2 0.00 0.00 0.0000 0.5846 0.000
g_qlogis 0.00 0.00 0.0000 0.0000 1.719
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2398 2390 -1179
Optimised parameters:
est. lower upper
cyan_0 100.8076 NA NA
log_k_JCZ38 -3.4684 NA NA
log_k_J9Z38 -5.0844 NA NA
log_k_JSE76 -5.5743 NA NA
f_cyan_ilr_1 0.6669 NA NA
f_cyan_ilr_2 0.7912 NA NA
f_JCZ38_qlogis 84.1825 NA NA
log_k1 -2.1671 NA NA
log_k2 -4.5447 NA NA
g_qlogis -0.5631 NA NA
a.1 2.9627 NA NA
b.1 0.0444 NA NA
SD.log_k_JCZ38 1.4044 NA NA
SD.log_k_J9Z38 0.6410 NA NA
SD.log_k_JSE76 0.5391 NA NA
SD.f_cyan_ilr_1 0.3203 NA NA
SD.f_cyan_ilr_2 0.5038 NA NA
SD.f_JCZ38_qlogis 3.5865 NA NA
SD.log_k2 0.3119 NA NA
SD.g_qlogis 0.8276 NA NA
Correlation is not available
Random effects:
est. lower upper
SD.log_k_JCZ38 1.4044 NA NA
SD.log_k_J9Z38 0.6410 NA NA
SD.log_k_JSE76 0.5391 NA NA
SD.f_cyan_ilr_1 0.3203 NA NA
SD.f_cyan_ilr_2 0.5038 NA NA
SD.f_JCZ38_qlogis 3.5865 NA NA
SD.log_k2 0.3119 NA NA
SD.g_qlogis 0.8276 NA NA
Variance model:
est. lower upper
a.1 2.9627 NA NA
b.1 0.0444 NA NA
Backtransformed parameters:
est. lower upper
cyan_0 1.008e+02 NA NA
k_JCZ38 3.117e-02 NA NA
k_J9Z38 6.193e-03 NA NA
k_JSE76 3.794e-03 NA NA
f_cyan_to_JCZ38 6.149e-01 NA NA
f_cyan_to_J9Z38 2.395e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
k1 1.145e-01 NA NA
k2 1.062e-02 NA NA
g 3.628e-01 NA NA
Resulting formation fractions:
ff
cyan_JCZ38 0.6149
cyan_J9Z38 0.2395
cyan_sink 0.1456
JCZ38_JSE76 1.0000
JCZ38_sink 0.0000
Estimated disappearance times:
DT50 DT90 DT50back DT50_k1 DT50_k2
cyan 26.26 174.32 52.47 6.053 65.25
JCZ38 22.24 73.88 NA NA NA
J9Z38 111.93 371.82 NA NA NA
JSE76 182.69 606.88 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:10:49 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan_free/dt = - k_cyan_free * cyan_free - k_cyan_free_bound *
cyan_free + k_cyan_bound_free * cyan_bound
d_cyan_bound/dt = + k_cyan_free_bound * cyan_free - k_cyan_bound_free *
cyan_bound
d_JCZ38/dt = + f_cyan_free_to_JCZ38 * k_cyan_free * cyan_free - k_JCZ38
* JCZ38
d_J9Z38/dt = + f_cyan_free_to_J9Z38 * k_cyan_free * cyan_free - k_J9Z38
* J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1279.472 s
Using 300, 100 iterations and 10 chains
Variance model: Constant variance
Starting values for degradation parameters:
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
102.0643 -2.8987 -2.7077
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38
-3.4717 -3.4008 -5.0024
log_k_JSE76 f_cyan_ilr_1 f_cyan_ilr_2
-5.8613 0.6855 1.2366
f_JCZ38_qlogis
13.7418
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
cyan_free_0 4.466 0.0000 0.000
log_k_cyan_free 0.000 0.6158 0.000
log_k_cyan_free_bound 0.000 0.0000 1.463
log_k_cyan_bound_free 0.000 0.0000 0.000
log_k_JCZ38 0.000 0.0000 0.000
log_k_J9Z38 0.000 0.0000 0.000
log_k_JSE76 0.000 0.0000 0.000
f_cyan_ilr_1 0.000 0.0000 0.000
f_cyan_ilr_2 0.000 0.0000 0.000
f_JCZ38_qlogis 0.000 0.0000 0.000
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38 log_k_JSE76
cyan_free_0 0.000 0.000 0.000 0.000
log_k_cyan_free 0.000 0.000 0.000 0.000
log_k_cyan_free_bound 0.000 0.000 0.000 0.000
log_k_cyan_bound_free 1.058 0.000 0.000 0.000
log_k_JCZ38 0.000 2.382 0.000 0.000
log_k_J9Z38 0.000 0.000 1.595 0.000
log_k_JSE76 0.000 0.000 0.000 1.245
f_cyan_ilr_1 0.000 0.000 0.000 0.000
f_cyan_ilr_2 0.000 0.000 0.000 0.000
f_JCZ38_qlogis 0.000 0.000 0.000 0.000
f_cyan_ilr_1 f_cyan_ilr_2 f_JCZ38_qlogis
cyan_free_0 0.0000 0.00 0.00
log_k_cyan_free 0.0000 0.00 0.00
log_k_cyan_free_bound 0.0000 0.00 0.00
log_k_cyan_bound_free 0.0000 0.00 0.00
log_k_JCZ38 0.0000 0.00 0.00
log_k_J9Z38 0.0000 0.00 0.00
log_k_JSE76 0.0000 0.00 0.00
f_cyan_ilr_1 0.6852 0.00 0.00
f_cyan_ilr_2 0.0000 1.28 0.00
f_JCZ38_qlogis 0.0000 0.00 16.14
Starting values for error model parameters:
a.1
1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2401 2394 -1181
Optimised parameters:
est. lower upper
cyan_free_0 102.7803 NA NA
log_k_cyan_free -2.8068 NA NA
log_k_cyan_free_bound -2.5714 NA NA
log_k_cyan_bound_free -3.4426 NA NA
log_k_JCZ38 -3.4994 NA NA
log_k_J9Z38 -5.1148 NA NA
log_k_JSE76 -5.6335 NA NA
f_cyan_ilr_1 0.6597 NA NA
f_cyan_ilr_2 0.5132 NA NA
f_JCZ38_qlogis 37.2090 NA NA
a.1 3.2367 NA NA
SD.log_k_cyan_free 0.3161 NA NA
SD.log_k_cyan_free_bound 0.8103 NA NA
SD.log_k_cyan_bound_free 0.5554 NA NA
SD.log_k_JCZ38 1.4858 NA NA
SD.log_k_J9Z38 0.5859 NA NA
SD.log_k_JSE76 0.6195 NA NA
SD.f_cyan_ilr_1 0.3118 NA NA
SD.f_cyan_ilr_2 0.3344 NA NA
SD.f_JCZ38_qlogis 0.5518 NA NA
Correlation is not available
Random effects:
est. lower upper
SD.log_k_cyan_free 0.3161 NA NA
SD.log_k_cyan_free_bound 0.8103 NA NA
SD.log_k_cyan_bound_free 0.5554 NA NA
SD.log_k_JCZ38 1.4858 NA NA
SD.log_k_J9Z38 0.5859 NA NA
SD.log_k_JSE76 0.6195 NA NA
SD.f_cyan_ilr_1 0.3118 NA NA
SD.f_cyan_ilr_2 0.3344 NA NA
SD.f_JCZ38_qlogis 0.5518 NA NA
Variance model:
est. lower upper
a.1 3.237 NA NA
Backtransformed parameters:
est. lower upper
cyan_free_0 1.028e+02 NA NA
k_cyan_free 6.040e-02 NA NA
k_cyan_free_bound 7.643e-02 NA NA
k_cyan_bound_free 3.198e-02 NA NA
k_JCZ38 3.022e-02 NA NA
k_J9Z38 6.007e-03 NA NA
k_JSE76 3.576e-03 NA NA
f_cyan_free_to_JCZ38 5.787e-01 NA NA
f_cyan_free_to_J9Z38 2.277e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
Estimated Eigenvalues of SFORB model(s):
cyan_b1 cyan_b2 cyan_g
0.15646 0.01235 0.33341
Resulting formation fractions:
ff
cyan_free_JCZ38 0.5787
cyan_free_J9Z38 0.2277
cyan_free_sink 0.1936
cyan_free 1.0000
JCZ38_JSE76 1.0000
JCZ38_sink 0.0000
Estimated disappearance times:
DT50 DT90 DT50back DT50_cyan_b1 DT50_cyan_b2
cyan 24.48 153.7 46.26 4.43 56.15
JCZ38 22.94 76.2 NA NA NA
J9Z38 115.39 383.3 NA NA NA
JSE76 193.84 643.9 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:17:00 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan_free/dt = - k_cyan_free * cyan_free - k_cyan_free_bound *
cyan_free + k_cyan_bound_free * cyan_bound
d_cyan_bound/dt = + k_cyan_free_bound * cyan_free - k_cyan_bound_free *
cyan_bound
d_JCZ38/dt = + f_cyan_free_to_JCZ38 * k_cyan_free * cyan_free - k_JCZ38
* JCZ38
d_J9Z38/dt = + f_cyan_free_to_J9Z38 * k_cyan_free * cyan_free - k_J9Z38
* J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1649.941 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
101.3964 -2.9881 -2.7949
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38
-3.4376 -3.3626 -4.9792
log_k_JSE76 f_cyan_ilr_1 f_cyan_ilr_2
-5.8727 0.6814 6.8139
f_JCZ38_qlogis
13.7419
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
cyan_free_0 5.317 0.0000 0.000
log_k_cyan_free 0.000 0.7301 0.000
log_k_cyan_free_bound 0.000 0.0000 1.384
log_k_cyan_bound_free 0.000 0.0000 0.000
log_k_JCZ38 0.000 0.0000 0.000
log_k_J9Z38 0.000 0.0000 0.000
log_k_JSE76 0.000 0.0000 0.000
f_cyan_ilr_1 0.000 0.0000 0.000
f_cyan_ilr_2 0.000 0.0000 0.000
f_JCZ38_qlogis 0.000 0.0000 0.000
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38 log_k_JSE76
cyan_free_0 0.000 0.000 0.000 0.000
log_k_cyan_free 0.000 0.000 0.000 0.000
log_k_cyan_free_bound 0.000 0.000 0.000 0.000
log_k_cyan_bound_free 1.109 0.000 0.000 0.000
log_k_JCZ38 0.000 2.272 0.000 0.000
log_k_J9Z38 0.000 0.000 1.633 0.000
log_k_JSE76 0.000 0.000 0.000 1.271
f_cyan_ilr_1 0.000 0.000 0.000 0.000
f_cyan_ilr_2 0.000 0.000 0.000 0.000
f_JCZ38_qlogis 0.000 0.000 0.000 0.000
f_cyan_ilr_1 f_cyan_ilr_2 f_JCZ38_qlogis
cyan_free_0 0.0000 0.00 0.00
log_k_cyan_free 0.0000 0.00 0.00
log_k_cyan_free_bound 0.0000 0.00 0.00
log_k_cyan_bound_free 0.0000 0.00 0.00
log_k_JCZ38 0.0000 0.00 0.00
log_k_J9Z38 0.0000 0.00 0.00
log_k_JSE76 0.0000 0.00 0.00
f_cyan_ilr_1 0.6838 0.00 0.00
f_cyan_ilr_2 0.0000 11.84 0.00
f_JCZ38_qlogis 0.0000 0.00 16.14
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2400 2392 -1180
Optimised parameters:
est. lower upper
cyan_free_0 100.69983 NA NA
log_k_cyan_free -3.11584 NA NA
log_k_cyan_free_bound -3.15216 NA NA
log_k_cyan_bound_free -3.65986 NA NA
log_k_JCZ38 -3.47811 NA NA
log_k_J9Z38 -5.08835 NA NA
log_k_JSE76 -5.55514 NA NA
f_cyan_ilr_1 0.66764 NA NA
f_cyan_ilr_2 0.78329 NA NA
f_JCZ38_qlogis 25.35245 NA NA
a.1 2.99088 NA NA
b.1 0.04346 NA NA
SD.log_k_cyan_free 0.48797 NA NA
SD.log_k_cyan_bound_free 0.27243 NA NA
SD.log_k_JCZ38 1.42450 NA NA
SD.log_k_J9Z38 0.63496 NA NA
SD.log_k_JSE76 0.55951 NA NA
SD.f_cyan_ilr_1 0.32687 NA NA
SD.f_cyan_ilr_2 0.48056 NA NA
SD.f_JCZ38_qlogis 0.43818 NA NA
Correlation is not available
Random effects:
est. lower upper
SD.log_k_cyan_free 0.4880 NA NA
SD.log_k_cyan_bound_free 0.2724 NA NA
SD.log_k_JCZ38 1.4245 NA NA
SD.log_k_J9Z38 0.6350 NA NA
SD.log_k_JSE76 0.5595 NA NA
SD.f_cyan_ilr_1 0.3269 NA NA
SD.f_cyan_ilr_2 0.4806 NA NA
SD.f_JCZ38_qlogis 0.4382 NA NA
Variance model:
est. lower upper
a.1 2.99088 NA NA
b.1 0.04346 NA NA
Backtransformed parameters:
est. lower upper
cyan_free_0 1.007e+02 NA NA
k_cyan_free 4.434e-02 NA NA
k_cyan_free_bound 4.276e-02 NA NA
k_cyan_bound_free 2.574e-02 NA NA
k_JCZ38 3.087e-02 NA NA
k_J9Z38 6.168e-03 NA NA
k_JSE76 3.868e-03 NA NA
f_cyan_free_to_JCZ38 6.143e-01 NA NA
f_cyan_free_to_J9Z38 2.389e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
Estimated Eigenvalues of SFORB model(s):
cyan_b1 cyan_b2 cyan_g
0.10161 0.01123 0.36636
Resulting formation fractions:
ff
cyan_free_JCZ38 6.143e-01
cyan_free_J9Z38 2.389e-01
cyan_free_sink 1.468e-01
cyan_free 1.000e+00
JCZ38_JSE76 1.000e+00
JCZ38_sink 9.763e-12
Estimated disappearance times:
DT50 DT90 DT50back DT50_cyan_b1 DT50_cyan_b2
cyan 25.91 164.4 49.49 6.822 61.72
JCZ38 22.46 74.6 NA NA NA
J9Z38 112.37 373.3 NA NA NA
JSE76 179.22 595.4 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:11:04 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - ifelse(time <= tb, k1, k2) * cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * ifelse(time <= tb, k1, k2) * cyan -
k_JCZ38 * JCZ38
d_J9Z38/dt = + f_cyan_to_J9Z38 * ifelse(time <= tb, k1, k2) * cyan -
k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1294.259 s
Using 300, 100 iterations and 10 chains
Variance model: Constant variance
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
102.8738 -3.4490 -4.9348 -5.5989 0.6469
f_cyan_ilr_2 f_JCZ38_qlogis log_k1 log_k2 log_tb
1.2854 9.7193 -2.9084 -4.1810 1.7813
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 5.409 0.00 0.00 0.000 0.0000
log_k_JCZ38 0.000 2.33 0.00 0.000 0.0000
log_k_J9Z38 0.000 0.00 1.59 0.000 0.0000
log_k_JSE76 0.000 0.00 0.00 1.006 0.0000
f_cyan_ilr_1 0.000 0.00 0.00 0.000 0.6371
f_cyan_ilr_2 0.000 0.00 0.00 0.000 0.0000
f_JCZ38_qlogis 0.000 0.00 0.00 0.000 0.0000
log_k1 0.000 0.00 0.00 0.000 0.0000
log_k2 0.000 0.00 0.00 0.000 0.0000
log_tb 0.000 0.00 0.00 0.000 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis log_k1 log_k2 log_tb
cyan_0 0.000 0.00 0.0000 0.0000 0.0000
log_k_JCZ38 0.000 0.00 0.0000 0.0000 0.0000
log_k_J9Z38 0.000 0.00 0.0000 0.0000 0.0000
log_k_JSE76 0.000 0.00 0.0000 0.0000 0.0000
f_cyan_ilr_1 0.000 0.00 0.0000 0.0000 0.0000
f_cyan_ilr_2 2.167 0.00 0.0000 0.0000 0.0000
f_JCZ38_qlogis 0.000 10.22 0.0000 0.0000 0.0000
log_k1 0.000 0.00 0.7003 0.0000 0.0000
log_k2 0.000 0.00 0.0000 0.8928 0.0000
log_tb 0.000 0.00 0.0000 0.0000 0.6774
Starting values for error model parameters:
a.1
1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2427 2420 -1194
Optimised parameters:
est. lower upper
cyan_0 101.84849 NA NA
log_k_JCZ38 -3.47365 NA NA
log_k_J9Z38 -5.10562 NA NA
log_k_JSE76 -5.60318 NA NA
f_cyan_ilr_1 0.66127 NA NA
f_cyan_ilr_2 0.60283 NA NA
f_JCZ38_qlogis 45.06408 NA NA
log_k1 -3.10124 NA NA
log_k2 -4.39028 NA NA
log_tb 2.32256 NA NA
a.1 3.32683 NA NA
SD.log_k_JCZ38 1.41427 NA NA
SD.log_k_J9Z38 0.54767 NA NA
SD.log_k_JSE76 0.62147 NA NA
SD.f_cyan_ilr_1 0.30189 NA NA
SD.f_cyan_ilr_2 0.34960 NA NA
SD.f_JCZ38_qlogis 0.04644 NA NA
SD.log_k1 0.39534 NA NA
SD.log_k2 0.43468 NA NA
SD.log_tb 0.60781 NA NA
Correlation is not available
Random effects:
est. lower upper
SD.log_k_JCZ38 1.41427 NA NA
SD.log_k_J9Z38 0.54767 NA NA
SD.log_k_JSE76 0.62147 NA NA
SD.f_cyan_ilr_1 0.30189 NA NA
SD.f_cyan_ilr_2 0.34960 NA NA
SD.f_JCZ38_qlogis 0.04644 NA NA
SD.log_k1 0.39534 NA NA
SD.log_k2 0.43468 NA NA
SD.log_tb 0.60781 NA NA
Variance model:
est. lower upper
a.1 3.327 NA NA
Backtransformed parameters:
est. lower upper
cyan_0 1.018e+02 NA NA
k_JCZ38 3.100e-02 NA NA
k_J9Z38 6.063e-03 NA NA
k_JSE76 3.686e-03 NA NA
f_cyan_to_JCZ38 5.910e-01 NA NA
f_cyan_to_J9Z38 2.320e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
k1 4.499e-02 NA NA
k2 1.240e-02 NA NA
tb 1.020e+01 NA NA
Resulting formation fractions:
ff
cyan_JCZ38 0.591
cyan_J9Z38 0.232
cyan_sink 0.177
JCZ38_JSE76 1.000
JCZ38_sink 0.000
Estimated disappearance times:
DT50 DT90 DT50back DT50_k1 DT50_k2
cyan 29.09 158.91 47.84 15.41 55.91
JCZ38 22.36 74.27 NA NA NA
J9Z38 114.33 379.80 NA NA NA
JSE76 188.04 624.66 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:11:24 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - ifelse(time <= tb, k1, k2) * cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * ifelse(time <= tb, k1, k2) * cyan -
k_JCZ38 * JCZ38
d_J9Z38/dt = + f_cyan_to_J9Z38 * ifelse(time <= tb, k1, k2) * cyan -
k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1313.805 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
101.168 -3.358 -4.941 -5.794 0.676
f_cyan_ilr_2 f_JCZ38_qlogis log_k1 log_k2 log_tb
5.740 13.863 -3.147 -4.262 2.173
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 5.79 0.000 0.000 0.000 0.0000
log_k_JCZ38 0.00 2.271 0.000 0.000 0.0000
log_k_J9Z38 0.00 0.000 1.614 0.000 0.0000
log_k_JSE76 0.00 0.000 0.000 1.264 0.0000
f_cyan_ilr_1 0.00 0.000 0.000 0.000 0.6761
f_cyan_ilr_2 0.00 0.000 0.000 0.000 0.0000
f_JCZ38_qlogis 0.00 0.000 0.000 0.000 0.0000
log_k1 0.00 0.000 0.000 0.000 0.0000
log_k2 0.00 0.000 0.000 0.000 0.0000
log_tb 0.00 0.000 0.000 0.000 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis log_k1 log_k2 log_tb
cyan_0 0.000 0.00 0.0000 0.0000 0.000
log_k_JCZ38 0.000 0.00 0.0000 0.0000 0.000
log_k_J9Z38 0.000 0.00 0.0000 0.0000 0.000
log_k_JSE76 0.000 0.00 0.0000 0.0000 0.000
f_cyan_ilr_1 0.000 0.00 0.0000 0.0000 0.000
f_cyan_ilr_2 9.572 0.00 0.0000 0.0000 0.000
f_JCZ38_qlogis 0.000 19.19 0.0000 0.0000 0.000
log_k1 0.000 0.00 0.8705 0.0000 0.000
log_k2 0.000 0.00 0.0000 0.9288 0.000
log_tb 0.000 0.00 0.0000 0.0000 1.065
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2422 2414 -1190
Optimised parameters:
est. lower upper
cyan_0 100.9521 NA NA
log_k_JCZ38 -3.4629 NA NA
log_k_J9Z38 -5.0346 NA NA
log_k_JSE76 -5.5722 NA NA
f_cyan_ilr_1 0.6560 NA NA
f_cyan_ilr_2 0.7983 NA NA
f_JCZ38_qlogis 42.7949 NA NA
log_k1 -3.1721 NA NA
log_k2 -4.4039 NA NA
log_tb 2.3994 NA NA
a.1 3.0586 NA NA
b.1 0.0380 NA NA
SD.log_k_JCZ38 1.3754 NA NA
SD.log_k_J9Z38 0.6703 NA NA
SD.log_k_JSE76 0.5876 NA NA
SD.f_cyan_ilr_1 0.3272 NA NA
SD.f_cyan_ilr_2 0.5300 NA NA
SD.f_JCZ38_qlogis 6.4465 NA NA
SD.log_k1 0.4135 NA NA
SD.log_k2 0.4182 NA NA
SD.log_tb 0.6035 NA NA
Correlation is not available
Random effects:
est. lower upper
SD.log_k_JCZ38 1.3754 NA NA
SD.log_k_J9Z38 0.6703 NA NA
SD.log_k_JSE76 0.5876 NA NA
SD.f_cyan_ilr_1 0.3272 NA NA
SD.f_cyan_ilr_2 0.5300 NA NA
SD.f_JCZ38_qlogis 6.4465 NA NA
SD.log_k1 0.4135 NA NA
SD.log_k2 0.4182 NA NA
SD.log_tb 0.6035 NA NA
Variance model:
est. lower upper
a.1 3.059 NA NA
b.1 0.038 NA NA
Backtransformed parameters:
est. lower upper
cyan_0 1.010e+02 NA NA
k_JCZ38 3.134e-02 NA NA
k_J9Z38 6.509e-03 NA NA
k_JSE76 3.802e-03 NA NA
f_cyan_to_JCZ38 6.127e-01 NA NA
f_cyan_to_J9Z38 2.423e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
k1 4.191e-02 NA NA
k2 1.223e-02 NA NA
tb 1.102e+01 NA NA
Resulting formation fractions:
ff
cyan_JCZ38 0.6127
cyan_J9Z38 0.2423
cyan_sink 0.1449
JCZ38_JSE76 1.0000
JCZ38_sink 0.0000
Estimated disappearance times:
DT50 DT90 DT50back DT50_k1 DT50_k2
cyan 29.94 161.54 48.63 16.54 56.68
JCZ38 22.12 73.47 NA NA NA
J9Z38 106.50 353.77 NA NA NA
JSE76 182.30 605.60 NA NA NA
Pathway 2
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:34:28 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - (alpha/beta) * 1/((time/beta) + 1) * cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * (alpha/beta) * 1/((time/beta) + 1) *
cyan - k_JCZ38 * JCZ38 + f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_to_J9Z38 * (alpha/beta) * 1/((time/beta) + 1) *
cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1030.246 s
Using 300, 100 iterations and 10 chains
Variance model: Constant variance
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
101.8173 -1.8998 -5.1449 -2.5415 0.6705
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_alpha log_beta
4.4669 16.1281 13.3327 -0.2314 2.8738
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 5.742 0.000 0.000 0.00 0.0000
log_k_JCZ38 0.000 1.402 0.000 0.00 0.0000
log_k_J9Z38 0.000 0.000 1.718 0.00 0.0000
log_k_JSE76 0.000 0.000 0.000 3.57 0.0000
f_cyan_ilr_1 0.000 0.000 0.000 0.00 0.5926
f_cyan_ilr_2 0.000 0.000 0.000 0.00 0.0000
f_JCZ38_qlogis 0.000 0.000 0.000 0.00 0.0000
f_JSE76_qlogis 0.000 0.000 0.000 0.00 0.0000
log_alpha 0.000 0.000 0.000 0.00 0.0000
log_beta 0.000 0.000 0.000 0.00 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_alpha log_beta
cyan_0 0.00 0.00 0.00 0.0000 0.0000
log_k_JCZ38 0.00 0.00 0.00 0.0000 0.0000
log_k_J9Z38 0.00 0.00 0.00 0.0000 0.0000
log_k_JSE76 0.00 0.00 0.00 0.0000 0.0000
f_cyan_ilr_1 0.00 0.00 0.00 0.0000 0.0000
f_cyan_ilr_2 10.56 0.00 0.00 0.0000 0.0000
f_JCZ38_qlogis 0.00 12.04 0.00 0.0000 0.0000
f_JSE76_qlogis 0.00 0.00 15.26 0.0000 0.0000
log_alpha 0.00 0.00 0.00 0.4708 0.0000
log_beta 0.00 0.00 0.00 0.0000 0.4432
Starting values for error model parameters:
a.1
1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2308 2301 -1134
Optimised parameters:
est. lower upper
cyan_0 101.9586 99.22024 104.69700
log_k_JCZ38 -2.4861 -3.17661 -1.79560
log_k_J9Z38 -5.3926 -6.08842 -4.69684
log_k_JSE76 -3.1193 -4.12904 -2.10962
f_cyan_ilr_1 0.7368 0.42085 1.05276
f_cyan_ilr_2 0.6196 0.06052 1.17861
f_JCZ38_qlogis 4.8970 -4.68003 14.47398
f_JSE76_qlogis 4.4066 -1.02087 9.83398
log_alpha -0.3021 -0.68264 0.07838
log_beta 2.7438 2.57970 2.90786
a.1 2.9008 2.69920 3.10245
SD.cyan_0 2.7081 0.64216 4.77401
SD.log_k_JCZ38 0.7043 0.19951 1.20907
SD.log_k_J9Z38 0.6248 0.05790 1.19180
SD.log_k_JSE76 1.0750 0.33157 1.81839
SD.f_cyan_ilr_1 0.3429 0.11688 0.56892
SD.f_cyan_ilr_2 0.4774 0.09381 0.86097
SD.f_JCZ38_qlogis 1.5565 -7.83970 10.95279
SD.f_JSE76_qlogis 1.6871 -1.25577 4.63000
SD.log_alpha 0.4216 0.15913 0.68405
Correlation:
cyan_0 l__JCZ3 l__J9Z3 l__JSE7 f_cy__1 f_cy__2 f_JCZ38 f_JSE76
log_k_JCZ38 -0.0167
log_k_J9Z38 -0.0307 0.0057
log_k_JSE76 -0.0032 0.1358 0.0009
f_cyan_ilr_1 -0.0087 0.0206 -0.1158 -0.0009
f_cyan_ilr_2 -0.1598 0.0690 0.1770 0.0002 -0.0007
f_JCZ38_qlogis 0.0966 -0.1132 -0.0440 0.0182 -0.1385 -0.4583
f_JSE76_qlogis -0.0647 0.1157 0.0333 -0.0026 0.1110 0.3620 -0.8586
log_alpha -0.0389 0.0113 0.0209 0.0021 0.0041 0.0451 -0.0605 0.0412
log_beta -0.2508 0.0533 0.0977 0.0098 0.0220 0.2741 -0.2934 0.1999
log_lph
log_k_JCZ38
log_k_J9Z38
log_k_JSE76
f_cyan_ilr_1
f_cyan_ilr_2
f_JCZ38_qlogis
f_JSE76_qlogis
log_alpha
log_beta 0.2281
Random effects:
est. lower upper
SD.cyan_0 2.7081 0.64216 4.7740
SD.log_k_JCZ38 0.7043 0.19951 1.2091
SD.log_k_J9Z38 0.6248 0.05790 1.1918
SD.log_k_JSE76 1.0750 0.33157 1.8184
SD.f_cyan_ilr_1 0.3429 0.11688 0.5689
SD.f_cyan_ilr_2 0.4774 0.09381 0.8610
SD.f_JCZ38_qlogis 1.5565 -7.83970 10.9528
SD.f_JSE76_qlogis 1.6871 -1.25577 4.6300
SD.log_alpha 0.4216 0.15913 0.6840
Variance model:
est. lower upper
a.1 2.901 2.699 3.102
Backtransformed parameters:
est. lower upper
cyan_0 101.95862 99.220240 1.047e+02
k_JCZ38 0.08323 0.041727 1.660e-01
k_J9Z38 0.00455 0.002269 9.124e-03
k_JSE76 0.04419 0.016098 1.213e-01
f_cyan_to_JCZ38 0.61318 NA NA
f_cyan_to_J9Z38 0.21630 NA NA
f_JCZ38_to_JSE76 0.99259 0.009193 1.000e+00
f_JSE76_to_JCZ38 0.98795 0.264857 9.999e-01
alpha 0.73924 0.505281 1.082e+00
beta 15.54568 13.193194 1.832e+01
Resulting formation fractions:
ff
cyan_JCZ38 0.613182
cyan_J9Z38 0.216298
cyan_sink 0.170519
JCZ38_JSE76 0.992586
JCZ38_sink 0.007414
JSE76_JCZ38 0.987950
JSE76_sink 0.012050
Estimated disappearance times:
DT50 DT90 DT50back
cyan 24.157 334.68 100.7
JCZ38 8.328 27.66 NA
J9Z38 152.341 506.06 NA
JSE76 15.687 52.11 NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:37:36 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - (alpha/beta) * 1/((time/beta) + 1) * cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * (alpha/beta) * 1/((time/beta) + 1) *
cyan - k_JCZ38 * JCZ38 + f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_to_J9Z38 * (alpha/beta) * 1/((time/beta) + 1) *
cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1217.619 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
101.9028 -1.9055 -5.0249 -2.5646 0.6807
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_alpha log_beta
4.8883 16.0676 9.3923 -0.1346 3.0364
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 6.321 0.000 0.000 0.000 0.0000
log_k_JCZ38 0.000 1.392 0.000 0.000 0.0000
log_k_J9Z38 0.000 0.000 1.561 0.000 0.0000
log_k_JSE76 0.000 0.000 0.000 3.614 0.0000
f_cyan_ilr_1 0.000 0.000 0.000 0.000 0.6339
f_cyan_ilr_2 0.000 0.000 0.000 0.000 0.0000
f_JCZ38_qlogis 0.000 0.000 0.000 0.000 0.0000
f_JSE76_qlogis 0.000 0.000 0.000 0.000 0.0000
log_alpha 0.000 0.000 0.000 0.000 0.0000
log_beta 0.000 0.000 0.000 0.000 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_alpha log_beta
cyan_0 0.00 0.00 0.00 0.0000 0.0000
log_k_JCZ38 0.00 0.00 0.00 0.0000 0.0000
log_k_J9Z38 0.00 0.00 0.00 0.0000 0.0000
log_k_JSE76 0.00 0.00 0.00 0.0000 0.0000
f_cyan_ilr_1 0.00 0.00 0.00 0.0000 0.0000
f_cyan_ilr_2 10.41 0.00 0.00 0.0000 0.0000
f_JCZ38_qlogis 0.00 12.24 0.00 0.0000 0.0000
f_JSE76_qlogis 0.00 0.00 15.13 0.0000 0.0000
log_alpha 0.00 0.00 0.00 0.3701 0.0000
log_beta 0.00 0.00 0.00 0.0000 0.5662
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2248 2240 -1103
Optimised parameters:
est. lower upper
cyan_0 101.55545 9.920e+01 1.039e+02
log_k_JCZ38 -2.37354 -2.928e+00 -1.819e+00
log_k_J9Z38 -5.14736 -5.960e+00 -4.335e+00
log_k_JSE76 -3.07802 -4.243e+00 -1.913e+00
f_cyan_ilr_1 0.71263 3.655e-01 1.060e+00
f_cyan_ilr_2 0.95202 2.701e-01 1.634e+00
f_JCZ38_qlogis 3.58473 1.251e+00 5.919e+00
f_JSE76_qlogis 19.03623 -1.037e+07 1.037e+07
log_alpha -0.15297 -4.490e-01 1.431e-01
log_beta 2.99230 2.706e+00 3.278e+00
a.1 2.04816 NA NA
b.1 0.06886 NA NA
SD.log_k_JCZ38 0.56174 NA NA
SD.log_k_J9Z38 0.86509 NA NA
SD.log_k_JSE76 1.28450 NA NA
SD.f_cyan_ilr_1 0.38705 NA NA
SD.f_cyan_ilr_2 0.54153 NA NA
SD.f_JCZ38_qlogis 1.65311 NA NA
SD.f_JSE76_qlogis 7.51468 NA NA
SD.log_alpha 0.31586 NA NA
SD.log_beta 0.24696 NA NA
Correlation is not available
Random effects:
est. lower upper
SD.log_k_JCZ38 0.5617 NA NA
SD.log_k_J9Z38 0.8651 NA NA
SD.log_k_JSE76 1.2845 NA NA
SD.f_cyan_ilr_1 0.3870 NA NA
SD.f_cyan_ilr_2 0.5415 NA NA
SD.f_JCZ38_qlogis 1.6531 NA NA
SD.f_JSE76_qlogis 7.5147 NA NA
SD.log_alpha 0.3159 NA NA
SD.log_beta 0.2470 NA NA
Variance model:
est. lower upper
a.1 2.04816 NA NA
b.1 0.06886 NA NA
Backtransformed parameters:
est. lower upper
cyan_0 1.016e+02 99.20301 103.9079
k_JCZ38 9.315e-02 0.05349 0.1622
k_J9Z38 5.815e-03 0.00258 0.0131
k_JSE76 4.605e-02 0.01436 0.1477
f_cyan_to_JCZ38 6.438e-01 NA NA
f_cyan_to_J9Z38 2.350e-01 NA NA
f_JCZ38_to_JSE76 9.730e-01 0.77745 0.9973
f_JSE76_to_JCZ38 1.000e+00 0.00000 1.0000
alpha 8.582e-01 0.63824 1.1538
beta 1.993e+01 14.97621 26.5262
Resulting formation fractions:
ff
cyan_JCZ38 6.438e-01
cyan_J9Z38 2.350e-01
cyan_sink 1.212e-01
JCZ38_JSE76 9.730e-01
JCZ38_sink 2.700e-02
JSE76_JCZ38 1.000e+00
JSE76_sink 5.403e-09
Estimated disappearance times:
DT50 DT90 DT50back
cyan 24.771 271.70 81.79
JCZ38 7.441 24.72 NA
J9Z38 119.205 395.99 NA
JSE76 15.052 50.00 NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:38:34 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 *
time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time)))
* cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_JCZ38 * JCZ38 +
f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_to_J9Z38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1276.128 s
Using 300, 100 iterations and 10 chains
Variance model: Constant variance
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
102.4358 -2.3107 -5.3123 -3.7120 0.6753
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_k1 log_k2
1.1462 12.4095 12.3630 -1.9317 -4.4557
g_qlogis
-0.5648
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 4.594 0.0000 0.000 0.0 0.0000
log_k_JCZ38 0.000 0.7966 0.000 0.0 0.0000
log_k_J9Z38 0.000 0.0000 1.561 0.0 0.0000
log_k_JSE76 0.000 0.0000 0.000 0.8 0.0000
f_cyan_ilr_1 0.000 0.0000 0.000 0.0 0.6349
f_cyan_ilr_2 0.000 0.0000 0.000 0.0 0.0000
f_JCZ38_qlogis 0.000 0.0000 0.000 0.0 0.0000
f_JSE76_qlogis 0.000 0.0000 0.000 0.0 0.0000
log_k1 0.000 0.0000 0.000 0.0 0.0000
log_k2 0.000 0.0000 0.000 0.0 0.0000
g_qlogis 0.000 0.0000 0.000 0.0 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_k1 log_k2
cyan_0 0.000 0.00 0.0 0.000 0.0000
log_k_JCZ38 0.000 0.00 0.0 0.000 0.0000
log_k_J9Z38 0.000 0.00 0.0 0.000 0.0000
log_k_JSE76 0.000 0.00 0.0 0.000 0.0000
f_cyan_ilr_1 0.000 0.00 0.0 0.000 0.0000
f_cyan_ilr_2 1.797 0.00 0.0 0.000 0.0000
f_JCZ38_qlogis 0.000 13.85 0.0 0.000 0.0000
f_JSE76_qlogis 0.000 0.00 14.1 0.000 0.0000
log_k1 0.000 0.00 0.0 1.106 0.0000
log_k2 0.000 0.00 0.0 0.000 0.6141
g_qlogis 0.000 0.00 0.0 0.000 0.0000
g_qlogis
cyan_0 0.000
log_k_JCZ38 0.000
log_k_J9Z38 0.000
log_k_JSE76 0.000
f_cyan_ilr_1 0.000
f_cyan_ilr_2 0.000
f_JCZ38_qlogis 0.000
f_JSE76_qlogis 0.000
log_k1 0.000
log_k2 0.000
g_qlogis 1.595
Starting values for error model parameters:
a.1
1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2290 2281 -1123
Optimised parameters:
est. lower upper
cyan_0 102.6903 101.44420 103.9365
log_k_JCZ38 -2.4018 -2.98058 -1.8230
log_k_J9Z38 -5.1865 -5.92931 -4.4437
log_k_JSE76 -3.0784 -4.25226 -1.9045
f_cyan_ilr_1 0.7157 0.37625 1.0551
f_cyan_ilr_2 0.7073 0.20136 1.2132
f_JCZ38_qlogis 4.6797 0.43240 8.9269
f_JSE76_qlogis 5.0080 -1.01380 11.0299
log_k1 -1.9620 -2.62909 -1.2949
log_k2 -4.4894 -4.94958 -4.0292
g_qlogis -0.4658 -1.34443 0.4129
a.1 2.7158 2.52576 2.9059
SD.log_k_JCZ38 0.5818 0.15679 1.0067
SD.log_k_J9Z38 0.7421 0.16751 1.3167
SD.log_k_JSE76 1.2841 0.43247 2.1356
SD.f_cyan_ilr_1 0.3748 0.13040 0.6192
SD.f_cyan_ilr_2 0.4550 0.08396 0.8261
SD.f_JCZ38_qlogis 2.0862 -0.73390 4.9062
SD.f_JSE76_qlogis 1.9585 -3.14773 7.0647
SD.log_k1 0.7389 0.25761 1.2201
SD.log_k2 0.5132 0.18143 0.8450
SD.g_qlogis 0.9870 0.35773 1.6164
Correlation:
cyan_0 l__JCZ3 l__J9Z3 l__JSE7 f_cy__1 f_cy__2 f_JCZ38 f_JSE76
log_k_JCZ38 -0.0170
log_k_J9Z38 -0.0457 0.0016
log_k_JSE76 -0.0046 0.1183 0.0005
f_cyan_ilr_1 0.0079 0.0072 -0.0909 0.0003
f_cyan_ilr_2 -0.3114 0.0343 0.1542 0.0023 -0.0519
f_JCZ38_qlogis 0.0777 -0.0601 -0.0152 0.0080 -0.0520 -0.2524
f_JSE76_qlogis -0.0356 0.0817 0.0073 0.0051 0.0388 0.1959 -0.6236
log_k1 0.0848 -0.0028 0.0010 -0.0010 -0.0014 -0.0245 0.0121 -0.0177
log_k2 0.0274 -0.0001 0.0075 0.0000 -0.0023 -0.0060 0.0000 -0.0130
g_qlogis 0.0159 0.0002 -0.0095 0.0002 0.0029 -0.0140 -0.0001 0.0149
log_k1 log_k2
log_k_JCZ38
log_k_J9Z38
log_k_JSE76
f_cyan_ilr_1
f_cyan_ilr_2
f_JCZ38_qlogis
f_JSE76_qlogis
log_k1
log_k2 0.0280
g_qlogis -0.0278 -0.0310
Random effects:
est. lower upper
SD.log_k_JCZ38 0.5818 0.15679 1.0067
SD.log_k_J9Z38 0.7421 0.16751 1.3167
SD.log_k_JSE76 1.2841 0.43247 2.1356
SD.f_cyan_ilr_1 0.3748 0.13040 0.6192
SD.f_cyan_ilr_2 0.4550 0.08396 0.8261
SD.f_JCZ38_qlogis 2.0862 -0.73390 4.9062
SD.f_JSE76_qlogis 1.9585 -3.14773 7.0647
SD.log_k1 0.7389 0.25761 1.2201
SD.log_k2 0.5132 0.18143 0.8450
SD.g_qlogis 0.9870 0.35773 1.6164
Variance model:
est. lower upper
a.1 2.716 2.526 2.906
Backtransformed parameters:
est. lower upper
cyan_0 1.027e+02 1.014e+02 103.93649
k_JCZ38 9.056e-02 5.076e-02 0.16154
k_J9Z38 5.591e-03 2.660e-03 0.01175
k_JSE76 4.603e-02 1.423e-02 0.14890
f_cyan_to_JCZ38 6.184e-01 NA NA
f_cyan_to_J9Z38 2.248e-01 NA NA
f_JCZ38_to_JSE76 9.908e-01 6.064e-01 0.99987
f_JSE76_to_JCZ38 9.934e-01 2.662e-01 0.99998
k1 1.406e-01 7.214e-02 0.27393
k2 1.123e-02 7.086e-03 0.01779
g 3.856e-01 2.068e-01 0.60177
Resulting formation fractions:
ff
cyan_JCZ38 0.618443
cyan_J9Z38 0.224770
cyan_sink 0.156787
JCZ38_JSE76 0.990803
JCZ38_sink 0.009197
JSE76_JCZ38 0.993360
JSE76_sink 0.006640
Estimated disappearance times:
DT50 DT90 DT50back DT50_k1 DT50_k2
cyan 21.674 161.70 48.68 4.931 61.74
JCZ38 7.654 25.43 NA NA NA
J9Z38 123.966 411.81 NA NA NA
JSE76 15.057 50.02 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:45:32 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 *
time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time)))
* cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_JCZ38 * JCZ38 +
f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_to_J9Z38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1693.767 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
101.7523 -1.5948 -5.0119 -2.2723 0.6719
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_k1 log_k2
5.1681 12.8238 12.4130 -2.0057 -4.5526
g_qlogis
-0.5805
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 5.627 0.000 0.000 0.000 0.0000
log_k_JCZ38 0.000 2.327 0.000 0.000 0.0000
log_k_J9Z38 0.000 0.000 1.664 0.000 0.0000
log_k_JSE76 0.000 0.000 0.000 4.566 0.0000
f_cyan_ilr_1 0.000 0.000 0.000 0.000 0.6519
f_cyan_ilr_2 0.000 0.000 0.000 0.000 0.0000
f_JCZ38_qlogis 0.000 0.000 0.000 0.000 0.0000
f_JSE76_qlogis 0.000 0.000 0.000 0.000 0.0000
log_k1 0.000 0.000 0.000 0.000 0.0000
log_k2 0.000 0.000 0.000 0.000 0.0000
g_qlogis 0.000 0.000 0.000 0.000 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_k1 log_k2
cyan_0 0.0 0.00 0.00 0.0000 0.0000
log_k_JCZ38 0.0 0.00 0.00 0.0000 0.0000
log_k_J9Z38 0.0 0.00 0.00 0.0000 0.0000
log_k_JSE76 0.0 0.00 0.00 0.0000 0.0000
f_cyan_ilr_1 0.0 0.00 0.00 0.0000 0.0000
f_cyan_ilr_2 10.1 0.00 0.00 0.0000 0.0000
f_JCZ38_qlogis 0.0 13.99 0.00 0.0000 0.0000
f_JSE76_qlogis 0.0 0.00 14.15 0.0000 0.0000
log_k1 0.0 0.00 0.00 0.8452 0.0000
log_k2 0.0 0.00 0.00 0.0000 0.5968
g_qlogis 0.0 0.00 0.00 0.0000 0.0000
g_qlogis
cyan_0 0.000
log_k_JCZ38 0.000
log_k_J9Z38 0.000
log_k_JSE76 0.000
f_cyan_ilr_1 0.000
f_cyan_ilr_2 0.000
f_JCZ38_qlogis 0.000
f_JSE76_qlogis 0.000
log_k1 0.000
log_k2 0.000
g_qlogis 1.691
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2234 2226 -1095
Optimised parameters:
est. lower upper
cyan_0 101.10667 9.903e+01 103.18265
log_k_JCZ38 -2.49437 -3.297e+00 -1.69221
log_k_J9Z38 -5.08171 -5.875e+00 -4.28846
log_k_JSE76 -3.20072 -4.180e+00 -2.22163
f_cyan_ilr_1 0.71059 3.639e-01 1.05727
f_cyan_ilr_2 1.15398 2.981e-01 2.00984
f_JCZ38_qlogis 3.18027 1.056e+00 5.30452
f_JSE76_qlogis 5.61578 -2.505e+01 36.28077
log_k1 -2.38875 -2.517e+00 -2.26045
log_k2 -4.67246 -4.928e+00 -4.41715
g_qlogis -0.28231 -1.135e+00 0.57058
a.1 2.08190 1.856e+00 2.30785
b.1 0.06114 5.015e-02 0.07214
SD.log_k_JCZ38 0.84622 2.637e-01 1.42873
SD.log_k_J9Z38 0.84564 2.566e-01 1.43464
SD.log_k_JSE76 1.04385 3.242e-01 1.76351
SD.f_cyan_ilr_1 0.38568 1.362e-01 0.63514
SD.f_cyan_ilr_2 0.68046 7.166e-02 1.28925
SD.f_JCZ38_qlogis 1.25244 -4.213e-02 2.54700
SD.f_JSE76_qlogis 0.28202 -1.515e+03 1515.87968
SD.log_k2 0.25749 7.655e-02 0.43843
SD.g_qlogis 0.94535 3.490e-01 1.54174
Correlation:
cyan_0 l__JCZ3 l__J9Z3 l__JSE7 f_cy__1 f_cy__2 f_JCZ38 f_JSE76
log_k_JCZ38 -0.0086
log_k_J9Z38 -0.0363 -0.0007
log_k_JSE76 0.0015 0.1210 -0.0017
f_cyan_ilr_1 -0.0048 0.0095 -0.0572 0.0030
f_cyan_ilr_2 -0.4788 0.0328 0.1143 0.0027 -0.0316
f_JCZ38_qlogis 0.0736 -0.0664 -0.0137 0.0145 -0.0444 -0.2175
f_JSE76_qlogis -0.0137 0.0971 0.0035 0.0009 0.0293 0.1333 -0.6767
log_k1 0.2345 -0.0350 -0.0099 -0.0113 -0.0126 -0.1652 0.1756 -0.2161
log_k2 0.0440 -0.0133 0.0199 -0.0040 -0.0097 -0.0119 0.0604 -0.1306
g_qlogis 0.0438 0.0078 -0.0123 0.0029 0.0046 -0.0363 -0.0318 0.0736
log_k1 log_k2
log_k_JCZ38
log_k_J9Z38
log_k_JSE76
f_cyan_ilr_1
f_cyan_ilr_2
f_JCZ38_qlogis
f_JSE76_qlogis
log_k1
log_k2 0.3198
g_qlogis -0.1666 -0.0954
Random effects:
est. lower upper
SD.log_k_JCZ38 0.8462 2.637e-01 1.4287
SD.log_k_J9Z38 0.8456 2.566e-01 1.4346
SD.log_k_JSE76 1.0439 3.242e-01 1.7635
SD.f_cyan_ilr_1 0.3857 1.362e-01 0.6351
SD.f_cyan_ilr_2 0.6805 7.166e-02 1.2893
SD.f_JCZ38_qlogis 1.2524 -4.213e-02 2.5470
SD.f_JSE76_qlogis 0.2820 -1.515e+03 1515.8797
SD.log_k2 0.2575 7.655e-02 0.4384
SD.g_qlogis 0.9453 3.490e-01 1.5417
Variance model:
est. lower upper
a.1 2.08190 1.85595 2.30785
b.1 0.06114 0.05015 0.07214
Backtransformed parameters:
est. lower upper
cyan_0 1.011e+02 9.903e+01 103.18265
k_JCZ38 8.255e-02 3.701e-02 0.18411
k_J9Z38 6.209e-03 2.809e-03 0.01373
k_JSE76 4.073e-02 1.530e-02 0.10843
f_cyan_to_JCZ38 6.608e-01 NA NA
f_cyan_to_J9Z38 2.419e-01 NA NA
f_JCZ38_to_JSE76 9.601e-01 7.419e-01 0.99506
f_JSE76_to_JCZ38 9.964e-01 1.322e-11 1.00000
k1 9.174e-02 8.070e-02 0.10430
k2 9.349e-03 7.243e-03 0.01207
g 4.299e-01 2.432e-01 0.63890
Resulting formation fractions:
ff
cyan_JCZ38 0.660808
cyan_J9Z38 0.241904
cyan_sink 0.097288
JCZ38_JSE76 0.960085
JCZ38_sink 0.039915
JSE76_JCZ38 0.996373
JSE76_sink 0.003627
Estimated disappearance times:
DT50 DT90 DT50back DT50_k1 DT50_k2
cyan 24.359 186.18 56.05 7.555 74.14
JCZ38 8.397 27.89 NA NA NA
J9Z38 111.631 370.83 NA NA NA
JSE76 17.017 56.53 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:38:37 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan_free/dt = - k_cyan_free * cyan_free - k_cyan_free_bound *
cyan_free + k_cyan_bound_free * cyan_bound
d_cyan_bound/dt = + k_cyan_free_bound * cyan_free - k_cyan_bound_free *
cyan_bound
d_JCZ38/dt = + f_cyan_free_to_JCZ38 * k_cyan_free * cyan_free - k_JCZ38
* JCZ38 + f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_free_to_J9Z38 * k_cyan_free * cyan_free - k_J9Z38
* J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1279.102 s
Using 300, 100 iterations and 10 chains
Variance model: Constant variance
Starting values for degradation parameters:
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
102.4394 -2.7673 -2.8942
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38
-3.6201 -2.3107 -5.3123
log_k_JSE76 f_cyan_ilr_1 f_cyan_ilr_2
-3.7120 0.6754 1.1448
f_JCZ38_qlogis f_JSE76_qlogis
13.2672 13.3538
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
cyan_free_0 4.589 0.0000 0.00
log_k_cyan_free 0.000 0.4849 0.00
log_k_cyan_free_bound 0.000 0.0000 1.62
log_k_cyan_bound_free 0.000 0.0000 0.00
log_k_JCZ38 0.000 0.0000 0.00
log_k_J9Z38 0.000 0.0000 0.00
log_k_JSE76 0.000 0.0000 0.00
f_cyan_ilr_1 0.000 0.0000 0.00
f_cyan_ilr_2 0.000 0.0000 0.00
f_JCZ38_qlogis 0.000 0.0000 0.00
f_JSE76_qlogis 0.000 0.0000 0.00
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38 log_k_JSE76
cyan_free_0 0.000 0.0000 0.000 0.0
log_k_cyan_free 0.000 0.0000 0.000 0.0
log_k_cyan_free_bound 0.000 0.0000 0.000 0.0
log_k_cyan_bound_free 1.197 0.0000 0.000 0.0
log_k_JCZ38 0.000 0.7966 0.000 0.0
log_k_J9Z38 0.000 0.0000 1.561 0.0
log_k_JSE76 0.000 0.0000 0.000 0.8
f_cyan_ilr_1 0.000 0.0000 0.000 0.0
f_cyan_ilr_2 0.000 0.0000 0.000 0.0
f_JCZ38_qlogis 0.000 0.0000 0.000 0.0
f_JSE76_qlogis 0.000 0.0000 0.000 0.0
f_cyan_ilr_1 f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis
cyan_free_0 0.0000 0.000 0.00 0.00
log_k_cyan_free 0.0000 0.000 0.00 0.00
log_k_cyan_free_bound 0.0000 0.000 0.00 0.00
log_k_cyan_bound_free 0.0000 0.000 0.00 0.00
log_k_JCZ38 0.0000 0.000 0.00 0.00
log_k_J9Z38 0.0000 0.000 0.00 0.00
log_k_JSE76 0.0000 0.000 0.00 0.00
f_cyan_ilr_1 0.6349 0.000 0.00 0.00
f_cyan_ilr_2 0.0000 1.797 0.00 0.00
f_JCZ38_qlogis 0.0000 0.000 13.84 0.00
f_JSE76_qlogis 0.0000 0.000 0.00 14.66
Starting values for error model parameters:
a.1
1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2284 2275 -1120
Optimised parameters:
est. lower upper
cyan_free_0 102.7730 1.015e+02 1.041e+02
log_k_cyan_free -2.8530 -3.167e+00 -2.539e+00
log_k_cyan_free_bound -2.7326 -3.543e+00 -1.922e+00
log_k_cyan_bound_free -3.5582 -4.126e+00 -2.990e+00
log_k_JCZ38 -2.3810 -2.921e+00 -1.841e+00
log_k_J9Z38 -5.2301 -5.963e+00 -4.497e+00
log_k_JSE76 -3.0286 -4.286e+00 -1.771e+00
f_cyan_ilr_1 0.7081 3.733e-01 1.043e+00
f_cyan_ilr_2 0.5847 7.846e-03 1.162e+00
f_JCZ38_qlogis 9.5676 -1.323e+03 1.342e+03
f_JSE76_qlogis 3.7042 7.254e-02 7.336e+00
a.1 2.7222 2.532e+00 2.913e+00
SD.log_k_cyan_free 0.3338 1.086e-01 5.589e-01
SD.log_k_cyan_free_bound 0.8888 3.023e-01 1.475e+00
SD.log_k_cyan_bound_free 0.6220 2.063e-01 1.038e+00
SD.log_k_JCZ38 0.5221 1.334e-01 9.108e-01
SD.log_k_J9Z38 0.7104 1.371e-01 1.284e+00
SD.log_k_JSE76 1.3837 4.753e-01 2.292e+00
SD.f_cyan_ilr_1 0.3620 1.248e-01 5.992e-01
SD.f_cyan_ilr_2 0.4259 8.145e-02 7.704e-01
SD.f_JCZ38_qlogis 3.5332 -1.037e+05 1.037e+05
SD.f_JSE76_qlogis 1.6990 -2.771e-01 3.675e+00
Correlation:
cyn_f_0 lg_k_c_ lg_k_cyn_f_ lg_k_cyn_b_ l__JCZ3 l__J9Z3
log_k_cyan_free 0.2126
log_k_cyan_free_bound 0.0894 0.0871
log_k_cyan_bound_free 0.0033 0.0410 0.0583
log_k_JCZ38 -0.0708 -0.0280 -0.0147 0.0019
log_k_J9Z38 -0.0535 -0.0138 0.0012 0.0148 0.0085
log_k_JSE76 -0.0066 -0.0030 -0.0021 -0.0005 0.1090 0.0010
f_cyan_ilr_1 -0.0364 -0.0157 -0.0095 -0.0015 0.0458 -0.0960
f_cyan_ilr_2 -0.3814 -0.1104 -0.0423 0.0146 0.1540 0.1526
f_JCZ38_qlogis 0.2507 0.0969 0.0482 -0.0097 -0.2282 -0.0363
f_JSE76_qlogis -0.1648 -0.0710 -0.0443 -0.0087 0.2002 0.0226
l__JSE7 f_cy__1 f_cy__2 f_JCZ38
log_k_cyan_free
log_k_cyan_free_bound
log_k_cyan_bound_free
log_k_JCZ38
log_k_J9Z38
log_k_JSE76
f_cyan_ilr_1 0.0001
f_cyan_ilr_2 0.0031 0.0586
f_JCZ38_qlogis 0.0023 -0.1867 -0.6255
f_JSE76_qlogis 0.0082 0.1356 0.4519 -0.7951
Random effects:
est. lower upper
SD.log_k_cyan_free 0.3338 1.086e-01 5.589e-01
SD.log_k_cyan_free_bound 0.8888 3.023e-01 1.475e+00
SD.log_k_cyan_bound_free 0.6220 2.063e-01 1.038e+00
SD.log_k_JCZ38 0.5221 1.334e-01 9.108e-01
SD.log_k_J9Z38 0.7104 1.371e-01 1.284e+00
SD.log_k_JSE76 1.3837 4.753e-01 2.292e+00
SD.f_cyan_ilr_1 0.3620 1.248e-01 5.992e-01
SD.f_cyan_ilr_2 0.4259 8.145e-02 7.704e-01
SD.f_JCZ38_qlogis 3.5332 -1.037e+05 1.037e+05
SD.f_JSE76_qlogis 1.6990 -2.771e-01 3.675e+00
Variance model:
est. lower upper
a.1 2.722 2.532 2.913
Backtransformed parameters:
est. lower upper
cyan_free_0 1.028e+02 1.015e+02 104.06475
k_cyan_free 5.767e-02 4.213e-02 0.07894
k_cyan_free_bound 6.505e-02 2.892e-02 0.14633
k_cyan_bound_free 2.849e-02 1.614e-02 0.05028
k_JCZ38 9.246e-02 5.390e-02 0.15859
k_J9Z38 5.353e-03 2.572e-03 0.01114
k_JSE76 4.838e-02 1.376e-02 0.17009
f_cyan_free_to_JCZ38 6.011e-01 5.028e-01 0.83792
f_cyan_free_to_J9Z38 2.208e-01 5.028e-01 0.83792
f_JCZ38_to_JSE76 9.999e-01 0.000e+00 1.00000
f_JSE76_to_JCZ38 9.760e-01 5.181e-01 0.99935
Estimated Eigenvalues of SFORB model(s):
cyan_b1 cyan_b2 cyan_g
0.13942 0.01178 0.35948
Resulting formation fractions:
ff
cyan_free_JCZ38 6.011e-01
cyan_free_J9Z38 2.208e-01
cyan_free_sink 1.780e-01
cyan_free 1.000e+00
JCZ38_JSE76 9.999e-01
JCZ38_sink 6.996e-05
JSE76_JCZ38 9.760e-01
JSE76_sink 2.403e-02
Estimated disappearance times:
DT50 DT90 DT50back DT50_cyan_b1 DT50_cyan_b2
cyan 23.390 157.60 47.44 4.971 58.82
JCZ38 7.497 24.90 NA NA NA
J9Z38 129.482 430.13 NA NA NA
JSE76 14.326 47.59 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 10:46:02 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan_free/dt = - k_cyan_free * cyan_free - k_cyan_free_bound *
cyan_free + k_cyan_bound_free * cyan_bound
d_cyan_bound/dt = + k_cyan_free_bound * cyan_free - k_cyan_bound_free *
cyan_bound
d_JCZ38/dt = + f_cyan_free_to_JCZ38 * k_cyan_free * cyan_free - k_JCZ38
* JCZ38 + f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_free_to_J9Z38 * k_cyan_free * cyan_free - k_J9Z38
* J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1723.343 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
101.751 -2.837 -3.016
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38
-3.660 -2.299 -5.313
log_k_JSE76 f_cyan_ilr_1 f_cyan_ilr_2
-3.699 0.672 5.873
f_JCZ38_qlogis f_JSE76_qlogis
13.216 13.338
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
cyan_free_0 5.629 0.000 0.000
log_k_cyan_free 0.000 0.446 0.000
log_k_cyan_free_bound 0.000 0.000 1.449
log_k_cyan_bound_free 0.000 0.000 0.000
log_k_JCZ38 0.000 0.000 0.000
log_k_J9Z38 0.000 0.000 0.000
log_k_JSE76 0.000 0.000 0.000
f_cyan_ilr_1 0.000 0.000 0.000
f_cyan_ilr_2 0.000 0.000 0.000
f_JCZ38_qlogis 0.000 0.000 0.000
f_JSE76_qlogis 0.000 0.000 0.000
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38 log_k_JSE76
cyan_free_0 0.000 0.0000 0.000 0.0000
log_k_cyan_free 0.000 0.0000 0.000 0.0000
log_k_cyan_free_bound 0.000 0.0000 0.000 0.0000
log_k_cyan_bound_free 1.213 0.0000 0.000 0.0000
log_k_JCZ38 0.000 0.7801 0.000 0.0000
log_k_J9Z38 0.000 0.0000 1.575 0.0000
log_k_JSE76 0.000 0.0000 0.000 0.8078
f_cyan_ilr_1 0.000 0.0000 0.000 0.0000
f_cyan_ilr_2 0.000 0.0000 0.000 0.0000
f_JCZ38_qlogis 0.000 0.0000 0.000 0.0000
f_JSE76_qlogis 0.000 0.0000 0.000 0.0000
f_cyan_ilr_1 f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis
cyan_free_0 0.0000 0.00 0.00 0.00
log_k_cyan_free 0.0000 0.00 0.00 0.00
log_k_cyan_free_bound 0.0000 0.00 0.00 0.00
log_k_cyan_bound_free 0.0000 0.00 0.00 0.00
log_k_JCZ38 0.0000 0.00 0.00 0.00
log_k_J9Z38 0.0000 0.00 0.00 0.00
log_k_JSE76 0.0000 0.00 0.00 0.00
f_cyan_ilr_1 0.6519 0.00 0.00 0.00
f_cyan_ilr_2 0.0000 10.78 0.00 0.00
f_JCZ38_qlogis 0.0000 0.00 13.96 0.00
f_JSE76_qlogis 0.0000 0.00 0.00 14.69
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2240 2232 -1098
Optimised parameters:
est. lower upper
cyan_free_0 101.10205 98.99221 103.2119
log_k_cyan_free -3.16929 -3.61395 -2.7246
log_k_cyan_free_bound -3.38259 -3.63022 -3.1350
log_k_cyan_bound_free -3.81075 -4.13888 -3.4826
log_k_JCZ38 -2.42057 -3.00756 -1.8336
log_k_J9Z38 -5.07501 -5.85138 -4.2986
log_k_JSE76 -3.12442 -4.21277 -2.0361
f_cyan_ilr_1 0.70577 0.35788 1.0537
f_cyan_ilr_2 1.14824 0.15810 2.1384
f_JCZ38_qlogis 3.52245 0.43257 6.6123
f_JSE76_qlogis 5.65140 -21.22295 32.5257
a.1 2.07062 1.84329 2.2980
b.1 0.06227 0.05124 0.0733
SD.log_k_cyan_free 0.49468 0.18566 0.8037
SD.log_k_cyan_bound_free 0.28972 0.07188 0.5076
SD.log_k_JCZ38 0.58852 0.16800 1.0090
SD.log_k_J9Z38 0.82500 0.24730 1.4027
SD.log_k_JSE76 1.19201 0.40313 1.9809
SD.f_cyan_ilr_1 0.38534 0.13640 0.6343
SD.f_cyan_ilr_2 0.72463 0.10076 1.3485
SD.f_JCZ38_qlogis 1.38223 -0.20997 2.9744
SD.f_JSE76_qlogis 2.07989 -72.53027 76.6901
Correlation:
cyn_f_0 lg_k_c_ lg_k_cyn_f_ lg_k_cyn_b_ l__JCZ3 l__J9Z3
log_k_cyan_free 0.1117
log_k_cyan_free_bound 0.1763 0.1828
log_k_cyan_bound_free 0.0120 0.0593 0.5030
log_k_JCZ38 -0.0459 -0.0230 -0.0931 -0.0337
log_k_J9Z38 -0.0381 -0.0123 -0.0139 0.0237 0.0063
log_k_JSE76 -0.0044 -0.0038 -0.0175 -0.0072 0.1120 0.0003
f_cyan_ilr_1 -0.0199 -0.0087 -0.0407 -0.0233 0.0268 -0.0552
f_cyan_ilr_2 -0.4806 -0.1015 -0.2291 -0.0269 0.1156 0.1113
f_JCZ38_qlogis 0.1805 0.0825 0.3085 0.0963 -0.1674 -0.0314
f_JSE76_qlogis -0.1586 -0.0810 -0.3560 -0.1563 0.2025 0.0278
l__JSE7 f_cy__1 f_cy__2 f_JCZ38
log_k_cyan_free
log_k_cyan_free_bound
log_k_cyan_bound_free
log_k_JCZ38
log_k_J9Z38
log_k_JSE76
f_cyan_ilr_1 0.0024
f_cyan_ilr_2 0.0087 0.0172
f_JCZ38_qlogis -0.0016 -0.1047 -0.4656
f_JSE76_qlogis 0.0119 0.1034 0.4584 -0.8137
Random effects:
est. lower upper
SD.log_k_cyan_free 0.4947 0.18566 0.8037
SD.log_k_cyan_bound_free 0.2897 0.07188 0.5076
SD.log_k_JCZ38 0.5885 0.16800 1.0090
SD.log_k_J9Z38 0.8250 0.24730 1.4027
SD.log_k_JSE76 1.1920 0.40313 1.9809
SD.f_cyan_ilr_1 0.3853 0.13640 0.6343
SD.f_cyan_ilr_2 0.7246 0.10076 1.3485
SD.f_JCZ38_qlogis 1.3822 -0.20997 2.9744
SD.f_JSE76_qlogis 2.0799 -72.53027 76.6901
Variance model:
est. lower upper
a.1 2.07062 1.84329 2.2980
b.1 0.06227 0.05124 0.0733
Backtransformed parameters:
est. lower upper
cyan_free_0 1.011e+02 9.899e+01 103.21190
k_cyan_free 4.203e-02 2.695e-02 0.06557
k_cyan_free_bound 3.396e-02 2.651e-02 0.04350
k_cyan_bound_free 2.213e-02 1.594e-02 0.03073
k_JCZ38 8.887e-02 4.941e-02 0.15984
k_J9Z38 6.251e-03 2.876e-03 0.01359
k_JSE76 4.396e-02 1.481e-02 0.13054
f_cyan_free_to_JCZ38 6.590e-01 5.557e-01 0.95365
f_cyan_free_to_J9Z38 2.429e-01 5.557e-01 0.95365
f_JCZ38_to_JSE76 9.713e-01 6.065e-01 0.99866
f_JSE76_to_JCZ38 9.965e-01 6.067e-10 1.00000
Estimated Eigenvalues of SFORB model(s):
cyan_b1 cyan_b2 cyan_g
0.08749 0.01063 0.40855
Resulting formation fractions:
ff
cyan_free_JCZ38 0.65905
cyan_free_J9Z38 0.24291
cyan_free_sink 0.09805
cyan_free 1.00000
JCZ38_JSE76 0.97132
JCZ38_sink 0.02868
JSE76_JCZ38 0.99650
JSE76_sink 0.00350
Estimated disappearance times:
DT50 DT90 DT50back DT50_cyan_b1 DT50_cyan_b2
cyan 24.91 167.16 50.32 7.922 65.19
JCZ38 7.80 25.91 NA NA NA
J9Z38 110.89 368.36 NA NA NA
JSE76 15.77 52.38 NA NA NA
Pathway 2, refined fits
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 11:18:41 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - (alpha/beta) * 1/((time/beta) + 1) * cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * (alpha/beta) * 1/((time/beta) + 1) *
cyan - k_JCZ38 * JCZ38 + f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_to_J9Z38 * (alpha/beta) * 1/((time/beta) + 1) *
cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1957.271 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
101.9028 -1.9055 -5.0249 -2.5646 0.6807
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_alpha log_beta
4.8883 16.0676 9.3923 -0.1346 3.0364
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 6.321 0.000 0.000 0.000 0.0000
log_k_JCZ38 0.000 1.392 0.000 0.000 0.0000
log_k_J9Z38 0.000 0.000 1.561 0.000 0.0000
log_k_JSE76 0.000 0.000 0.000 3.614 0.0000
f_cyan_ilr_1 0.000 0.000 0.000 0.000 0.6339
f_cyan_ilr_2 0.000 0.000 0.000 0.000 0.0000
f_JCZ38_qlogis 0.000 0.000 0.000 0.000 0.0000
f_JSE76_qlogis 0.000 0.000 0.000 0.000 0.0000
log_alpha 0.000 0.000 0.000 0.000 0.0000
log_beta 0.000 0.000 0.000 0.000 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_alpha log_beta
cyan_0 0.00 0.00 0.00 0.0000 0.0000
log_k_JCZ38 0.00 0.00 0.00 0.0000 0.0000
log_k_J9Z38 0.00 0.00 0.00 0.0000 0.0000
log_k_JSE76 0.00 0.00 0.00 0.0000 0.0000
f_cyan_ilr_1 0.00 0.00 0.00 0.0000 0.0000
f_cyan_ilr_2 10.41 0.00 0.00 0.0000 0.0000
f_JCZ38_qlogis 0.00 12.24 0.00 0.0000 0.0000
f_JSE76_qlogis 0.00 0.00 15.13 0.0000 0.0000
log_alpha 0.00 0.00 0.00 0.3701 0.0000
log_beta 0.00 0.00 0.00 0.0000 0.5662
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2251 2244 -1106
Optimised parameters:
est. lower upper
cyan_0 101.05768 NA NA
log_k_JCZ38 -2.73252 NA NA
log_k_J9Z38 -5.07399 NA NA
log_k_JSE76 -3.52863 NA NA
f_cyan_ilr_1 0.72176 NA NA
f_cyan_ilr_2 1.34610 NA NA
f_JCZ38_qlogis 2.08337 NA NA
f_JSE76_qlogis 1590.31880 NA NA
log_alpha -0.09336 NA NA
log_beta 3.10191 NA NA
a.1 2.08557 1.85439 2.31675
b.1 0.06998 0.05800 0.08197
SD.log_k_JCZ38 1.20053 0.43329 1.96777
SD.log_k_J9Z38 0.85854 0.26708 1.45000
SD.log_k_JSE76 0.62528 0.16061 1.08995
SD.f_cyan_ilr_1 0.35190 0.12340 0.58039
SD.f_cyan_ilr_2 0.85385 0.15391 1.55378
SD.log_alpha 0.28971 0.08718 0.49225
SD.log_beta 0.31614 0.05938 0.57290
Correlation is not available
Random effects:
est. lower upper
SD.log_k_JCZ38 1.2005 0.43329 1.9678
SD.log_k_J9Z38 0.8585 0.26708 1.4500
SD.log_k_JSE76 0.6253 0.16061 1.0900
SD.f_cyan_ilr_1 0.3519 0.12340 0.5804
SD.f_cyan_ilr_2 0.8538 0.15391 1.5538
SD.log_alpha 0.2897 0.08718 0.4923
SD.log_beta 0.3161 0.05938 0.5729
Variance model:
est. lower upper
a.1 2.08557 1.854 2.31675
b.1 0.06998 0.058 0.08197
Backtransformed parameters:
est. lower upper
cyan_0 1.011e+02 NA NA
k_JCZ38 6.506e-02 NA NA
k_J9Z38 6.257e-03 NA NA
k_JSE76 2.935e-02 NA NA
f_cyan_to_JCZ38 6.776e-01 NA NA
f_cyan_to_J9Z38 2.442e-01 NA NA
f_JCZ38_to_JSE76 8.893e-01 NA NA
f_JSE76_to_JCZ38 1.000e+00 NA NA
alpha 9.109e-01 NA NA
beta 2.224e+01 NA NA
Resulting formation fractions:
ff
cyan_JCZ38 0.67761
cyan_J9Z38 0.24417
cyan_sink 0.07822
JCZ38_JSE76 0.88928
JCZ38_sink 0.11072
JSE76_JCZ38 1.00000
JSE76_sink 0.00000
Estimated disappearance times:
DT50 DT90 DT50back
cyan 25.36 256.37 77.18
JCZ38 10.65 35.39 NA
J9Z38 110.77 367.98 NA
JSE76 23.62 78.47 NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 11:16:32 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 *
time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time)))
* cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_JCZ38 * JCZ38 +
f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_to_J9Z38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1828.403 s
Using 300, 100 iterations and 10 chains
Variance model: Constant variance
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
102.4358 -2.3107 -5.3123 -3.7120 0.6753
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_k1 log_k2
1.1462 12.4095 12.3630 -1.9317 -4.4557
g_qlogis
-0.5648
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 4.594 0.0000 0.000 0.0 0.0000
log_k_JCZ38 0.000 0.7966 0.000 0.0 0.0000
log_k_J9Z38 0.000 0.0000 1.561 0.0 0.0000
log_k_JSE76 0.000 0.0000 0.000 0.8 0.0000
f_cyan_ilr_1 0.000 0.0000 0.000 0.0 0.6349
f_cyan_ilr_2 0.000 0.0000 0.000 0.0 0.0000
f_JCZ38_qlogis 0.000 0.0000 0.000 0.0 0.0000
f_JSE76_qlogis 0.000 0.0000 0.000 0.0 0.0000
log_k1 0.000 0.0000 0.000 0.0 0.0000
log_k2 0.000 0.0000 0.000 0.0 0.0000
g_qlogis 0.000 0.0000 0.000 0.0 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_k1 log_k2
cyan_0 0.000 0.00 0.0 0.000 0.0000
log_k_JCZ38 0.000 0.00 0.0 0.000 0.0000
log_k_J9Z38 0.000 0.00 0.0 0.000 0.0000
log_k_JSE76 0.000 0.00 0.0 0.000 0.0000
f_cyan_ilr_1 0.000 0.00 0.0 0.000 0.0000
f_cyan_ilr_2 1.797 0.00 0.0 0.000 0.0000
f_JCZ38_qlogis 0.000 13.85 0.0 0.000 0.0000
f_JSE76_qlogis 0.000 0.00 14.1 0.000 0.0000
log_k1 0.000 0.00 0.0 1.106 0.0000
log_k2 0.000 0.00 0.0 0.000 0.6141
g_qlogis 0.000 0.00 0.0 0.000 0.0000
g_qlogis
cyan_0 0.000
log_k_JCZ38 0.000
log_k_J9Z38 0.000
log_k_JSE76 0.000
f_cyan_ilr_1 0.000
f_cyan_ilr_2 0.000
f_JCZ38_qlogis 0.000
f_JSE76_qlogis 0.000
log_k1 0.000
log_k2 0.000
g_qlogis 1.595
Starting values for error model parameters:
a.1
1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2282 2274 -1121
Optimised parameters:
est. lower upper
cyan_0 102.5254 NA NA
log_k_JCZ38 -2.9358 NA NA
log_k_J9Z38 -5.1424 NA NA
log_k_JSE76 -3.6458 NA NA
f_cyan_ilr_1 0.6957 NA NA
f_cyan_ilr_2 0.6635 NA NA
f_JCZ38_qlogis 4984.8163 NA NA
f_JSE76_qlogis 1.9415 NA NA
log_k1 -1.9456 NA NA
log_k2 -4.4705 NA NA
g_qlogis -0.5117 NA NA
a.1 2.7455 2.55392 2.9370
SD.log_k_JCZ38 1.3163 0.47635 2.1563
SD.log_k_J9Z38 0.7162 0.16133 1.2711
SD.log_k_JSE76 0.6457 0.15249 1.1390
SD.f_cyan_ilr_1 0.3424 0.11714 0.5677
SD.f_cyan_ilr_2 0.4524 0.09709 0.8077
SD.log_k1 0.7353 0.25445 1.2161
SD.log_k2 0.5137 0.18206 0.8453
SD.g_qlogis 0.9857 0.35651 1.6148
Correlation is not available
Random effects:
est. lower upper
SD.log_k_JCZ38 1.3163 0.47635 2.1563
SD.log_k_J9Z38 0.7162 0.16133 1.2711
SD.log_k_JSE76 0.6457 0.15249 1.1390
SD.f_cyan_ilr_1 0.3424 0.11714 0.5677
SD.f_cyan_ilr_2 0.4524 0.09709 0.8077
SD.log_k1 0.7353 0.25445 1.2161
SD.log_k2 0.5137 0.18206 0.8453
SD.g_qlogis 0.9857 0.35651 1.6148
Variance model:
est. lower upper
a.1 2.745 2.554 2.937
Backtransformed parameters:
est. lower upper
cyan_0 1.025e+02 NA NA
k_JCZ38 5.309e-02 NA NA
k_J9Z38 5.844e-03 NA NA
k_JSE76 2.610e-02 NA NA
f_cyan_to_JCZ38 6.079e-01 NA NA
f_cyan_to_J9Z38 2.272e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
f_JSE76_to_JCZ38 8.745e-01 NA NA
k1 1.429e-01 NA NA
k2 1.144e-02 NA NA
g 3.748e-01 NA NA
Resulting formation fractions:
ff
cyan_JCZ38 0.6079
cyan_J9Z38 0.2272
cyan_sink 0.1649
JCZ38_JSE76 1.0000
JCZ38_sink 0.0000
JSE76_JCZ38 0.8745
JSE76_sink 0.1255
Estimated disappearance times:
DT50 DT90 DT50back DT50_k1 DT50_k2
cyan 22.29 160.20 48.22 4.85 60.58
JCZ38 13.06 43.37 NA NA NA
J9Z38 118.61 394.02 NA NA NA
JSE76 26.56 88.22 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 11:22:28 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 *
time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time)))
* cyan
d_JCZ38/dt = + f_cyan_to_JCZ38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_JCZ38 * JCZ38 +
f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_to_J9Z38 * ((k1 * g * exp(-k1 * time) + k2 * (1 -
g) * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
exp(-k2 * time))) * cyan - k_J9Z38 * J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 2183.989 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
101.7523 -1.5948 -5.0119 -2.2723 0.6719
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_k1 log_k2
5.1681 12.8238 12.4130 -2.0057 -4.5526
g_qlogis
-0.5805
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_0 log_k_JCZ38 log_k_J9Z38 log_k_JSE76 f_cyan_ilr_1
cyan_0 5.627 0.000 0.000 0.000 0.0000
log_k_JCZ38 0.000 2.327 0.000 0.000 0.0000
log_k_J9Z38 0.000 0.000 1.664 0.000 0.0000
log_k_JSE76 0.000 0.000 0.000 4.566 0.0000
f_cyan_ilr_1 0.000 0.000 0.000 0.000 0.6519
f_cyan_ilr_2 0.000 0.000 0.000 0.000 0.0000
f_JCZ38_qlogis 0.000 0.000 0.000 0.000 0.0000
f_JSE76_qlogis 0.000 0.000 0.000 0.000 0.0000
log_k1 0.000 0.000 0.000 0.000 0.0000
log_k2 0.000 0.000 0.000 0.000 0.0000
g_qlogis 0.000 0.000 0.000 0.000 0.0000
f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis log_k1 log_k2
cyan_0 0.0 0.00 0.00 0.0000 0.0000
log_k_JCZ38 0.0 0.00 0.00 0.0000 0.0000
log_k_J9Z38 0.0 0.00 0.00 0.0000 0.0000
log_k_JSE76 0.0 0.00 0.00 0.0000 0.0000
f_cyan_ilr_1 0.0 0.00 0.00 0.0000 0.0000
f_cyan_ilr_2 10.1 0.00 0.00 0.0000 0.0000
f_JCZ38_qlogis 0.0 13.99 0.00 0.0000 0.0000
f_JSE76_qlogis 0.0 0.00 14.15 0.0000 0.0000
log_k1 0.0 0.00 0.00 0.8452 0.0000
log_k2 0.0 0.00 0.00 0.0000 0.5968
g_qlogis 0.0 0.00 0.00 0.0000 0.0000
g_qlogis
cyan_0 0.000
log_k_JCZ38 0.000
log_k_J9Z38 0.000
log_k_JSE76 0.000
f_cyan_ilr_1 0.000
f_cyan_ilr_2 0.000
f_JCZ38_qlogis 0.000
f_JSE76_qlogis 0.000
log_k1 0.000
log_k2 0.000
g_qlogis 1.691
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2232 2224 -1096
Optimised parameters:
est. lower upper
cyan_0 101.20051 NA NA
log_k_JCZ38 -2.93542 NA NA
log_k_J9Z38 -5.03151 NA NA
log_k_JSE76 -3.67679 NA NA
f_cyan_ilr_1 0.67290 NA NA
f_cyan_ilr_2 0.99787 NA NA
f_JCZ38_qlogis 348.32484 NA NA
f_JSE76_qlogis 1.87846 NA NA
log_k1 -2.32738 NA NA
log_k2 -4.61295 NA NA
g_qlogis -0.38342 NA NA
a.1 2.06184 1.83746 2.28622
b.1 0.06329 0.05211 0.07447
SD.log_k_JCZ38 1.29042 0.47468 2.10617
SD.log_k_J9Z38 0.84235 0.25903 1.42566
SD.log_k_JSE76 0.56930 0.13934 0.99926
SD.f_cyan_ilr_1 0.35183 0.12298 0.58068
SD.f_cyan_ilr_2 0.77269 0.17908 1.36631
SD.log_k2 0.28549 0.09210 0.47888
SD.g_qlogis 0.93830 0.34568 1.53093
Correlation is not available
Random effects:
est. lower upper
SD.log_k_JCZ38 1.2904 0.4747 2.1062
SD.log_k_J9Z38 0.8423 0.2590 1.4257
SD.log_k_JSE76 0.5693 0.1393 0.9993
SD.f_cyan_ilr_1 0.3518 0.1230 0.5807
SD.f_cyan_ilr_2 0.7727 0.1791 1.3663
SD.log_k2 0.2855 0.0921 0.4789
SD.g_qlogis 0.9383 0.3457 1.5309
Variance model:
est. lower upper
a.1 2.06184 1.83746 2.28622
b.1 0.06329 0.05211 0.07447
Backtransformed parameters:
est. lower upper
cyan_0 1.012e+02 NA NA
k_JCZ38 5.311e-02 NA NA
k_J9Z38 6.529e-03 NA NA
k_JSE76 2.530e-02 NA NA
f_cyan_to_JCZ38 6.373e-01 NA NA
f_cyan_to_J9Z38 2.461e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
f_JSE76_to_JCZ38 8.674e-01 NA NA
k1 9.755e-02 NA NA
k2 9.922e-03 NA NA
g 4.053e-01 NA NA
Resulting formation fractions:
ff
cyan_JCZ38 0.6373
cyan_J9Z38 0.2461
cyan_sink 0.1167
JCZ38_JSE76 1.0000
JCZ38_sink 0.0000
JSE76_JCZ38 0.8674
JSE76_sink 0.1326
Estimated disappearance times:
DT50 DT90 DT50back DT50_k1 DT50_k2
cyan 24.93 179.68 54.09 7.105 69.86
JCZ38 13.05 43.36 NA NA NA
J9Z38 106.16 352.67 NA NA NA
JSE76 27.39 91.00 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 11:17:37 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan_free/dt = - k_cyan_free * cyan_free - k_cyan_free_bound *
cyan_free + k_cyan_bound_free * cyan_bound
d_cyan_bound/dt = + k_cyan_free_bound * cyan_free - k_cyan_bound_free *
cyan_bound
d_JCZ38/dt = + f_cyan_free_to_JCZ38 * k_cyan_free * cyan_free - k_JCZ38
* JCZ38 + f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_free_to_J9Z38 * k_cyan_free * cyan_free - k_J9Z38
* J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 1893.29 s
Using 300, 100 iterations and 10 chains
Variance model: Constant variance
Starting values for degradation parameters:
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
102.4394 -2.7673 -2.8942
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38
-3.6201 -2.3107 -5.3123
log_k_JSE76 f_cyan_ilr_1 f_cyan_ilr_2
-3.7120 0.6754 1.1448
f_JCZ38_qlogis f_JSE76_qlogis
13.2672 13.3538
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
cyan_free_0 4.589 0.0000 0.00
log_k_cyan_free 0.000 0.4849 0.00
log_k_cyan_free_bound 0.000 0.0000 1.62
log_k_cyan_bound_free 0.000 0.0000 0.00
log_k_JCZ38 0.000 0.0000 0.00
log_k_J9Z38 0.000 0.0000 0.00
log_k_JSE76 0.000 0.0000 0.00
f_cyan_ilr_1 0.000 0.0000 0.00
f_cyan_ilr_2 0.000 0.0000 0.00
f_JCZ38_qlogis 0.000 0.0000 0.00
f_JSE76_qlogis 0.000 0.0000 0.00
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38 log_k_JSE76
cyan_free_0 0.000 0.0000 0.000 0.0
log_k_cyan_free 0.000 0.0000 0.000 0.0
log_k_cyan_free_bound 0.000 0.0000 0.000 0.0
log_k_cyan_bound_free 1.197 0.0000 0.000 0.0
log_k_JCZ38 0.000 0.7966 0.000 0.0
log_k_J9Z38 0.000 0.0000 1.561 0.0
log_k_JSE76 0.000 0.0000 0.000 0.8
f_cyan_ilr_1 0.000 0.0000 0.000 0.0
f_cyan_ilr_2 0.000 0.0000 0.000 0.0
f_JCZ38_qlogis 0.000 0.0000 0.000 0.0
f_JSE76_qlogis 0.000 0.0000 0.000 0.0
f_cyan_ilr_1 f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis
cyan_free_0 0.0000 0.000 0.00 0.00
log_k_cyan_free 0.0000 0.000 0.00 0.00
log_k_cyan_free_bound 0.0000 0.000 0.00 0.00
log_k_cyan_bound_free 0.0000 0.000 0.00 0.00
log_k_JCZ38 0.0000 0.000 0.00 0.00
log_k_J9Z38 0.0000 0.000 0.00 0.00
log_k_JSE76 0.0000 0.000 0.00 0.00
f_cyan_ilr_1 0.6349 0.000 0.00 0.00
f_cyan_ilr_2 0.0000 1.797 0.00 0.00
f_JCZ38_qlogis 0.0000 0.000 13.84 0.00
f_JSE76_qlogis 0.0000 0.000 0.00 14.66
Starting values for error model parameters:
a.1
1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2279 2272 -1120
Optimised parameters:
est. lower upper
cyan_free_0 102.5621 NA NA
log_k_cyan_free -2.8531 NA NA
log_k_cyan_free_bound -2.6916 NA NA
log_k_cyan_bound_free -3.5032 NA NA
log_k_JCZ38 -2.9436 NA NA
log_k_J9Z38 -5.1140 NA NA
log_k_JSE76 -3.6472 NA NA
f_cyan_ilr_1 0.6887 NA NA
f_cyan_ilr_2 0.6874 NA NA
f_JCZ38_qlogis 4063.6389 NA NA
f_JSE76_qlogis 1.9556 NA NA
a.1 2.7460 2.55451 2.9376
SD.log_k_cyan_free 0.3131 0.09841 0.5277
SD.log_k_cyan_free_bound 0.8850 0.29909 1.4710
SD.log_k_cyan_bound_free 0.6167 0.20391 1.0295
SD.log_k_JCZ38 1.3555 0.49101 2.2200
SD.log_k_J9Z38 0.7200 0.16166 1.2783
SD.log_k_JSE76 0.6252 0.14619 1.1042
SD.f_cyan_ilr_1 0.3386 0.11447 0.5627
SD.f_cyan_ilr_2 0.4699 0.09810 0.8417
Correlation is not available
Random effects:
est. lower upper
SD.log_k_cyan_free 0.3131 0.09841 0.5277
SD.log_k_cyan_free_bound 0.8850 0.29909 1.4710
SD.log_k_cyan_bound_free 0.6167 0.20391 1.0295
SD.log_k_JCZ38 1.3555 0.49101 2.2200
SD.log_k_J9Z38 0.7200 0.16166 1.2783
SD.log_k_JSE76 0.6252 0.14619 1.1042
SD.f_cyan_ilr_1 0.3386 0.11447 0.5627
SD.f_cyan_ilr_2 0.4699 0.09810 0.8417
Variance model:
est. lower upper
a.1 2.746 2.555 2.938
Backtransformed parameters:
est. lower upper
cyan_free_0 1.026e+02 NA NA
k_cyan_free 5.767e-02 NA NA
k_cyan_free_bound 6.777e-02 NA NA
k_cyan_bound_free 3.010e-02 NA NA
k_JCZ38 5.267e-02 NA NA
k_J9Z38 6.012e-03 NA NA
k_JSE76 2.606e-02 NA NA
f_cyan_free_to_JCZ38 6.089e-01 NA NA
f_cyan_free_to_J9Z38 2.299e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
f_JSE76_to_JCZ38 8.761e-01 NA NA
Estimated Eigenvalues of SFORB model(s):
cyan_b1 cyan_b2 cyan_g
0.1434 0.0121 0.3469
Resulting formation fractions:
ff
cyan_free_JCZ38 0.6089
cyan_free_J9Z38 0.2299
cyan_free_sink 0.1612
cyan_free 1.0000
JCZ38_JSE76 1.0000
JCZ38_sink 0.0000
JSE76_JCZ38 0.8761
JSE76_sink 0.1239
Estimated disappearance times:
DT50 DT90 DT50back DT50_cyan_b1 DT50_cyan_b2
cyan 23.94 155.06 46.68 4.832 57.28
JCZ38 13.16 43.71 NA NA NA
J9Z38 115.30 383.02 NA NA NA
JSE76 26.59 88.35 NA NA NA
saemix version used for fitting: 3.2
mkin version used for pre-fitting: 1.2.2
R version used for fitting: 4.2.2
Date of fit: Sat Jan 28 11:21:01 2023
Date of summary: Fri Feb 17 22:24:33 2023
Equations:
d_cyan_free/dt = - k_cyan_free * cyan_free - k_cyan_free_bound *
cyan_free + k_cyan_bound_free * cyan_bound
d_cyan_bound/dt = + k_cyan_free_bound * cyan_free - k_cyan_bound_free *
cyan_bound
d_JCZ38/dt = + f_cyan_free_to_JCZ38 * k_cyan_free * cyan_free - k_JCZ38
* JCZ38 + f_JSE76_to_JCZ38 * k_JSE76 * JSE76
d_J9Z38/dt = + f_cyan_free_to_J9Z38 * k_cyan_free * cyan_free - k_J9Z38
* J9Z38
d_JSE76/dt = + f_JCZ38_to_JSE76 * k_JCZ38 * JCZ38 - k_JSE76 * JSE76
Data:
433 observations of 4 variable(s) grouped in 5 datasets
Model predictions using solution type deSolve
Fitted in 2097.842 s
Using 300, 100 iterations and 10 chains
Variance model: Two-component variance function
Starting values for degradation parameters:
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
101.751 -2.837 -3.016
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38
-3.660 -2.299 -5.313
log_k_JSE76 f_cyan_ilr_1 f_cyan_ilr_2
-3.699 0.672 5.873
f_JCZ38_qlogis f_JSE76_qlogis
13.216 13.338
Fixed degradation parameter values:
None
Starting values for random effects (square root of initial entries in omega):
cyan_free_0 log_k_cyan_free log_k_cyan_free_bound
cyan_free_0 5.629 0.000 0.000
log_k_cyan_free 0.000 0.446 0.000
log_k_cyan_free_bound 0.000 0.000 1.449
log_k_cyan_bound_free 0.000 0.000 0.000
log_k_JCZ38 0.000 0.000 0.000
log_k_J9Z38 0.000 0.000 0.000
log_k_JSE76 0.000 0.000 0.000
f_cyan_ilr_1 0.000 0.000 0.000
f_cyan_ilr_2 0.000 0.000 0.000
f_JCZ38_qlogis 0.000 0.000 0.000
f_JSE76_qlogis 0.000 0.000 0.000
log_k_cyan_bound_free log_k_JCZ38 log_k_J9Z38 log_k_JSE76
cyan_free_0 0.000 0.0000 0.000 0.0000
log_k_cyan_free 0.000 0.0000 0.000 0.0000
log_k_cyan_free_bound 0.000 0.0000 0.000 0.0000
log_k_cyan_bound_free 1.213 0.0000 0.000 0.0000
log_k_JCZ38 0.000 0.7801 0.000 0.0000
log_k_J9Z38 0.000 0.0000 1.575 0.0000
log_k_JSE76 0.000 0.0000 0.000 0.8078
f_cyan_ilr_1 0.000 0.0000 0.000 0.0000
f_cyan_ilr_2 0.000 0.0000 0.000 0.0000
f_JCZ38_qlogis 0.000 0.0000 0.000 0.0000
f_JSE76_qlogis 0.000 0.0000 0.000 0.0000
f_cyan_ilr_1 f_cyan_ilr_2 f_JCZ38_qlogis f_JSE76_qlogis
cyan_free_0 0.0000 0.00 0.00 0.00
log_k_cyan_free 0.0000 0.00 0.00 0.00
log_k_cyan_free_bound 0.0000 0.00 0.00 0.00
log_k_cyan_bound_free 0.0000 0.00 0.00 0.00
log_k_JCZ38 0.0000 0.00 0.00 0.00
log_k_J9Z38 0.0000 0.00 0.00 0.00
log_k_JSE76 0.0000 0.00 0.00 0.00
f_cyan_ilr_1 0.6519 0.00 0.00 0.00
f_cyan_ilr_2 0.0000 10.78 0.00 0.00
f_JCZ38_qlogis 0.0000 0.00 13.96 0.00
f_JSE76_qlogis 0.0000 0.00 0.00 14.69
Starting values for error model parameters:
a.1 b.1
1 1
Results:
Likelihood computed by importance sampling
AIC BIC logLik
2236 2228 -1098
Optimised parameters:
est. lower upper
cyan_free_0 100.72760 NA NA
log_k_cyan_free -3.18281 NA NA
log_k_cyan_free_bound -3.37924 NA NA
log_k_cyan_bound_free -3.77107 NA NA
log_k_JCZ38 -2.92811 NA NA
log_k_J9Z38 -5.02759 NA NA
log_k_JSE76 -3.65835 NA NA
f_cyan_ilr_1 0.67390 NA NA
f_cyan_ilr_2 1.15106 NA NA
f_JCZ38_qlogis 827.82299 NA NA
f_JSE76_qlogis 1.83064 NA NA
a.1 2.06921 1.84443 2.29399
b.1 0.06391 0.05267 0.07515
SD.log_k_cyan_free 0.50518 0.18962 0.82075
SD.log_k_cyan_bound_free 0.30991 0.08170 0.53813
SD.log_k_JCZ38 1.26661 0.46578 2.06744
SD.log_k_J9Z38 0.88272 0.27813 1.48730
SD.log_k_JSE76 0.53050 0.12561 0.93538
SD.f_cyan_ilr_1 0.35547 0.12461 0.58633
SD.f_cyan_ilr_2 0.91446 0.20131 1.62761
Correlation is not available
Random effects:
est. lower upper
SD.log_k_cyan_free 0.5052 0.1896 0.8207
SD.log_k_cyan_bound_free 0.3099 0.0817 0.5381
SD.log_k_JCZ38 1.2666 0.4658 2.0674
SD.log_k_J9Z38 0.8827 0.2781 1.4873
SD.log_k_JSE76 0.5305 0.1256 0.9354
SD.f_cyan_ilr_1 0.3555 0.1246 0.5863
SD.f_cyan_ilr_2 0.9145 0.2013 1.6276
Variance model:
est. lower upper
a.1 2.06921 1.84443 2.29399
b.1 0.06391 0.05267 0.07515
Backtransformed parameters:
est. lower upper
cyan_free_0 1.007e+02 NA NA
k_cyan_free 4.147e-02 NA NA
k_cyan_free_bound 3.407e-02 NA NA
k_cyan_bound_free 2.303e-02 NA NA
k_JCZ38 5.350e-02 NA NA
k_J9Z38 6.555e-03 NA NA
k_JSE76 2.578e-02 NA NA
f_cyan_free_to_JCZ38 6.505e-01 NA NA
f_cyan_free_to_J9Z38 2.508e-01 NA NA
f_JCZ38_to_JSE76 1.000e+00 NA NA
f_JSE76_to_JCZ38 8.618e-01 NA NA
Estimated Eigenvalues of SFORB model(s):
cyan_b1 cyan_b2 cyan_g
0.08768 0.01089 0.39821
Resulting formation fractions:
ff
cyan_free_JCZ38 0.65053
cyan_free_J9Z38 0.25082
cyan_free_sink 0.09864
cyan_free 1.00000
JCZ38_JSE76 1.00000
JCZ38_sink 0.00000
JSE76_JCZ38 0.86184
JSE76_sink 0.13816
Estimated disappearance times:
DT50 DT90 DT50back DT50_cyan_b1 DT50_cyan_b2
cyan 25.32 164.79 49.61 7.906 63.64
JCZ38 12.96 43.04 NA NA NA
J9Z38 105.75 351.29 NA NA NA
JSE76 26.89 89.33 NA NA NA
Session info
R version 4.2.2 Patched (2022-11-10 r83330)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Debian GNU/Linux bookworm/sid
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-serial/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-serial/libopenblas-r0.3.21.so
locale:
[1] LC_CTYPE=de_DE.UTF-8 LC_NUMERIC=C
[3] LC_TIME=de_DE.UTF-8 LC_COLLATE=de_DE.UTF-8
[5] LC_MONETARY=de_DE.UTF-8 LC_MESSAGES=de_DE.UTF-8
[7] LC_PAPER=de_DE.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=de_DE.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] saemix_3.2 npde_3.3 knitr_1.41 mkin_1.2.3
loaded via a namespace (and not attached):
[1] pillar_1.8.1 bslib_0.4.2 compiler_4.2.2 jquerylib_0.1.4
[5] tools_4.2.2 mclust_6.0.0 digest_0.6.31 tibble_3.1.8
[9] jsonlite_1.8.4 evaluate_0.19 memoise_2.0.1 lifecycle_1.0.3
[13] nlme_3.1-162 gtable_0.3.1 lattice_0.20-45 pkgconfig_2.0.3
[17] rlang_1.0.6 DBI_1.1.3 cli_3.5.0 yaml_2.3.6
[21] pkgdown_2.0.7 xfun_0.35 fastmap_1.1.0 gridExtra_2.3
[25] dplyr_1.0.10 stringr_1.5.0 generics_0.1.3 desc_1.4.2
[29] fs_1.5.2 vctrs_0.5.1 sass_0.4.4 systemfonts_1.0.4
[33] tidyselect_1.2.0 rprojroot_2.0.3 lmtest_0.9-40 grid_4.2.2
[37] inline_0.3.19 glue_1.6.2 R6_2.5.1 textshaping_0.3.6
[41] fansi_1.0.3 rmarkdown_2.19 purrr_1.0.0 ggplot2_3.4.0
[45] magrittr_2.0.3 scales_1.2.1 htmltools_0.5.4 assertthat_0.2.1
[49] colorspace_2.0-3 ragg_1.2.4 utf8_1.2.2 stringi_1.7.8
[53] munsell_0.5.0 cachem_1.0.6 zoo_1.8-11