The class is initialised with an identifier which is generally an ISO common name. Additional chemical information is retrieved from the internet if available.
Format
An R6Class generator object
Super class
chents::chent -> pai
Public fields
isoISO common name according to ISO 1750 as retreived from pesticidecompendium.bcpc.org
bcpcList of information retrieved from pesticidecompendium.bcpc.org
Methods
Inherited methods
chents::chent$add_PUF()chents::chent$add_TP()chents::chent$add_cwsat()chents::chent$add_p0()chents::chent$add_soil_degradation()chents::chent$add_soil_ff()chents::chent$add_soil_sorption()chents::chent$add_transformation()chents::chent$emf()chents::chent$get_chyaml()chents::chent$get_pubchem()chents::chent$get_rdkit()chents::chent$pdf()chents::chent$png()chents::chent$try_pubchem()
Method new()
Usage
pai$new(
iso,
identifier = iso,
smiles = NULL,
inchikey = NULL,
bcpc = TRUE,
pubchem = TRUE,
pubchem_from = "auto",
rdkit = TRUE,
template = NULL,
chyaml = TRUE
)Examples
# On Travis, we get a certificate validation error,
# likely because the system (xenial) is so old,
# therefore don't run this example on Travis
if (Sys.getenv("TRAVIS") == "") {
atr <- pai$new("atrazine")
print(atr)
if (!is.null(atr$Picture)) {
plot(atr)
}
}
#> BCPC:
#> PubChem:
#> Trying to get chemical information from RDKit using PubChem_Canonical SMILES
#> CCNC1=NC(=NC(=N1)Cl)NC(C)C
#> Did not find chyaml file ./atrazine.yaml
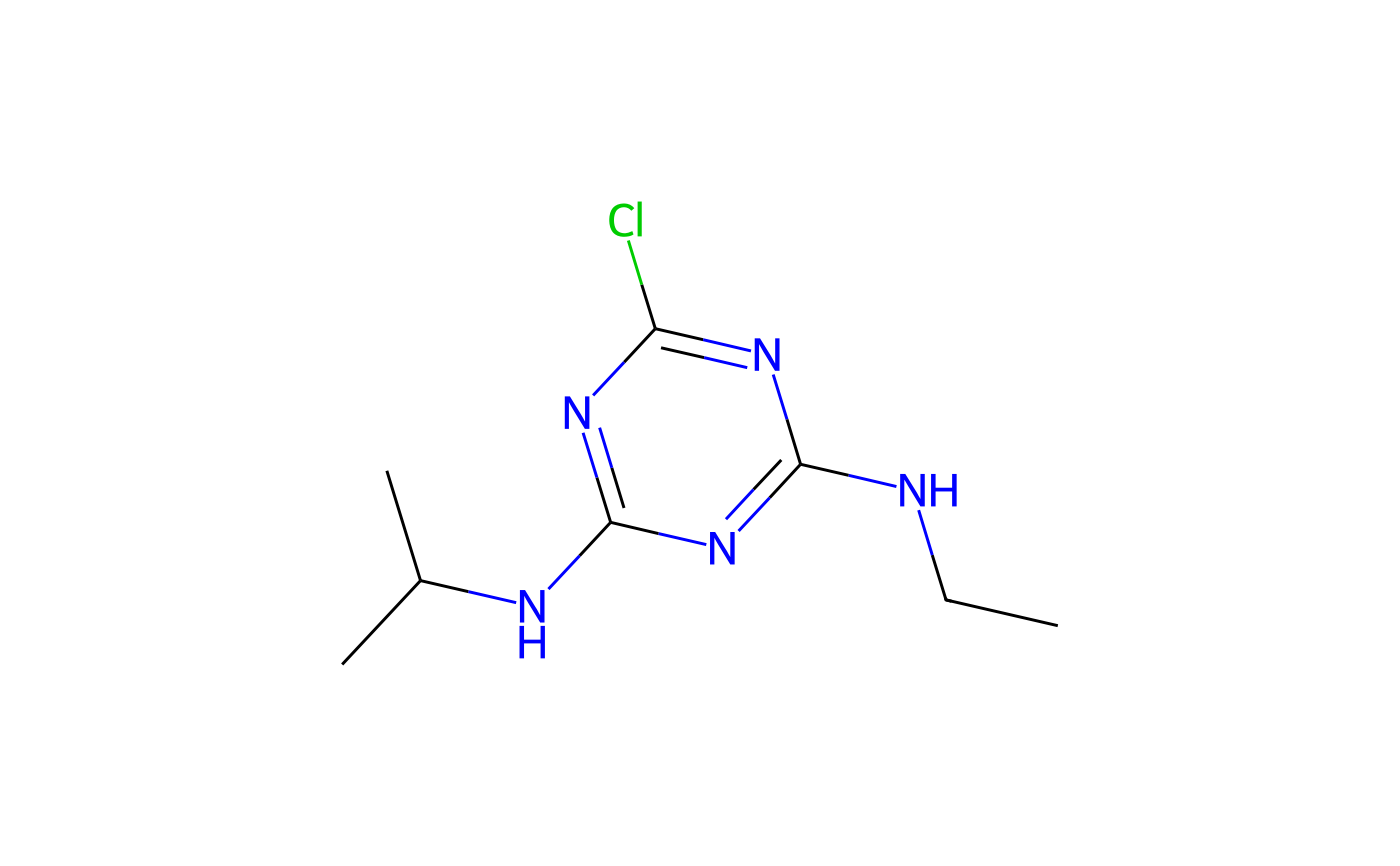 #> <pai> with ISO common name $iso atrazine
#> <chent>
#> Identifier $identifier atrazine
#> InChI Key $inchikey MXWJVTOOROXGIU-UHFFFAOYSA-N
#> SMILES string $smiles:
#> PubChem_Canonical
#> "CCNC1=NC(=NC(=N1)Cl)NC(C)C"
#> Molecular weight $mw: 215.7
#> PubChem synonyms (up to 10):
#> [1] "atrazine" "1912-24-9" "Gesaprim" "Oleogesaprim" "Aktikon"
#> [6] "Atranex" "Chromozin" "Atazinax" "Atrasine" "Gesoprim"
#> <pai> with ISO common name $iso atrazine
#> <chent>
#> Identifier $identifier atrazine
#> InChI Key $inchikey MXWJVTOOROXGIU-UHFFFAOYSA-N
#> SMILES string $smiles:
#> PubChem_Canonical
#> "CCNC1=NC(=NC(=N1)Cl)NC(C)C"
#> Molecular weight $mw: 215.7
#> PubChem synonyms (up to 10):
#> [1] "atrazine" "1912-24-9" "Gesaprim" "Oleogesaprim" "Aktikon"
#> [6] "Atranex" "Chromozin" "Atazinax" "Atrasine" "Gesoprim"