This function uses saemix::saemix() as a backend for fitting nonlinear mixed
effects models created from mmkin row objects using the Stochastic Approximation
Expectation Maximisation algorithm (SAEM).
saem(object, control, ...) # S3 method for mmkin saem( object, control = list(displayProgress = FALSE, print = FALSE, save = FALSE, save.graphs = FALSE), cores = 1, verbose = FALSE, suppressPlot = TRUE, quiet = FALSE, ... ) # S3 method for saem.mmkin print(x, digits = max(3, getOption("digits") - 3), ...) saemix_model(object, cores = 1, verbose = FALSE, ...) saemix_data(object, verbose = FALSE, ...)
Arguments
| object | An mmkin row object containing several fits of the same mkinmod model to different datasets |
|---|---|
| control | Passed to saemix::saemix |
| ... | Further parameters passed to saemix::saemixModel. |
| cores | The number of cores to be used for multicore processing using
|
| verbose | Should we print information about created objects of type saemix::SaemixModel and saemix::SaemixData? |
| suppressPlot | Should we suppress any plotting that is done by the saemix function? |
| quiet | Should we suppress the messages saemix prints at the beginning and the end of the optimisation process? |
| x | An saem.mmkin object to print |
| digits | Number of digits to use for printing |
Value
An S3 object of class 'saem.mmkin', containing the fitted saemix::SaemixObject as a list component named 'so'. The object also inherits from 'mixed.mmkin'.
An saemix::SaemixModel object.
An saemix::SaemixData object.
Details
An mmkin row object is essentially a list of mkinfit objects that have been obtained by fitting the same model to a list of datasets using mkinfit.
Starting values for the fixed effects (population mean parameters, argument
psi0 of saemix::saemixModel() are the mean values of the parameters found
using mmkin.
See also
Examples
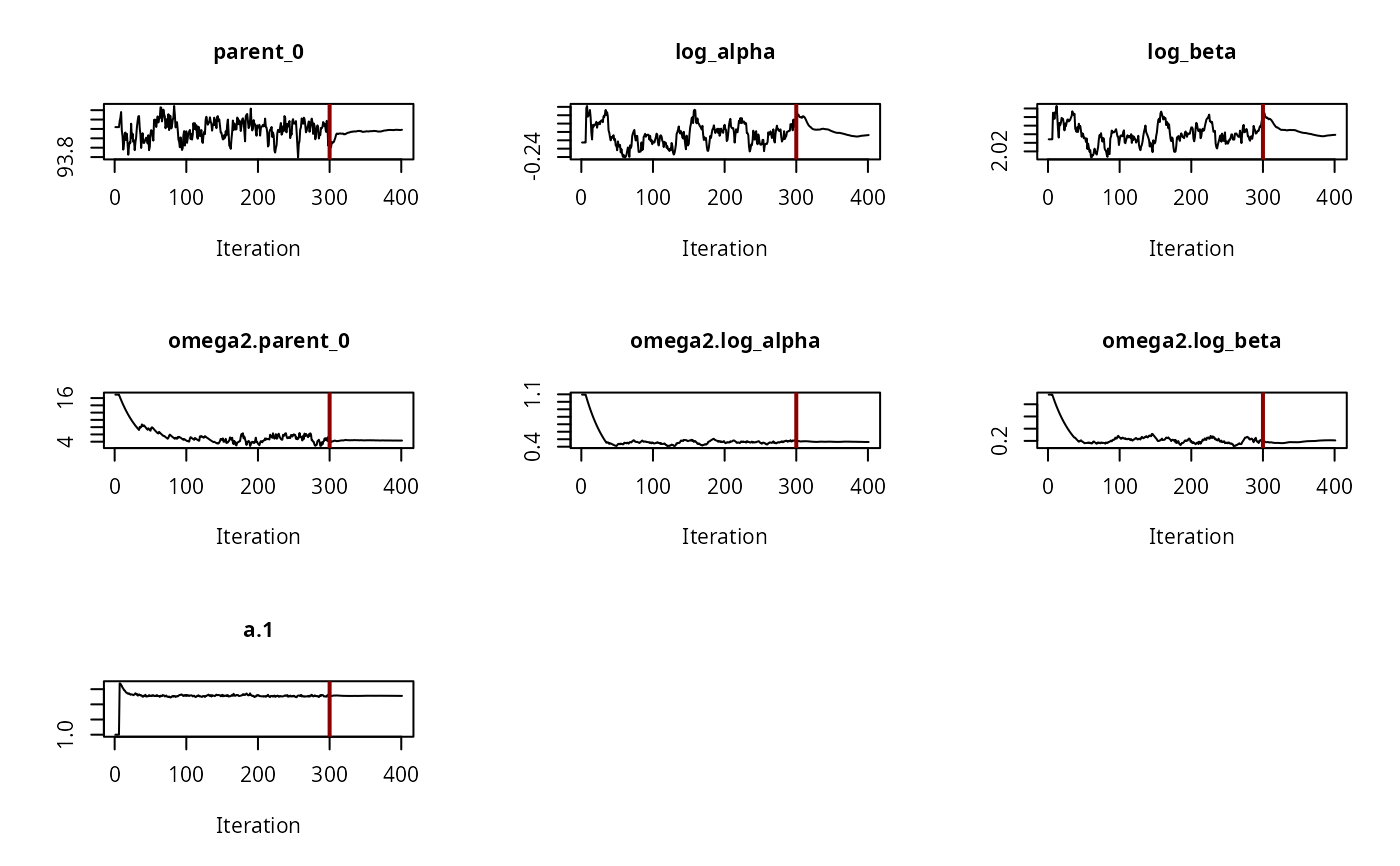
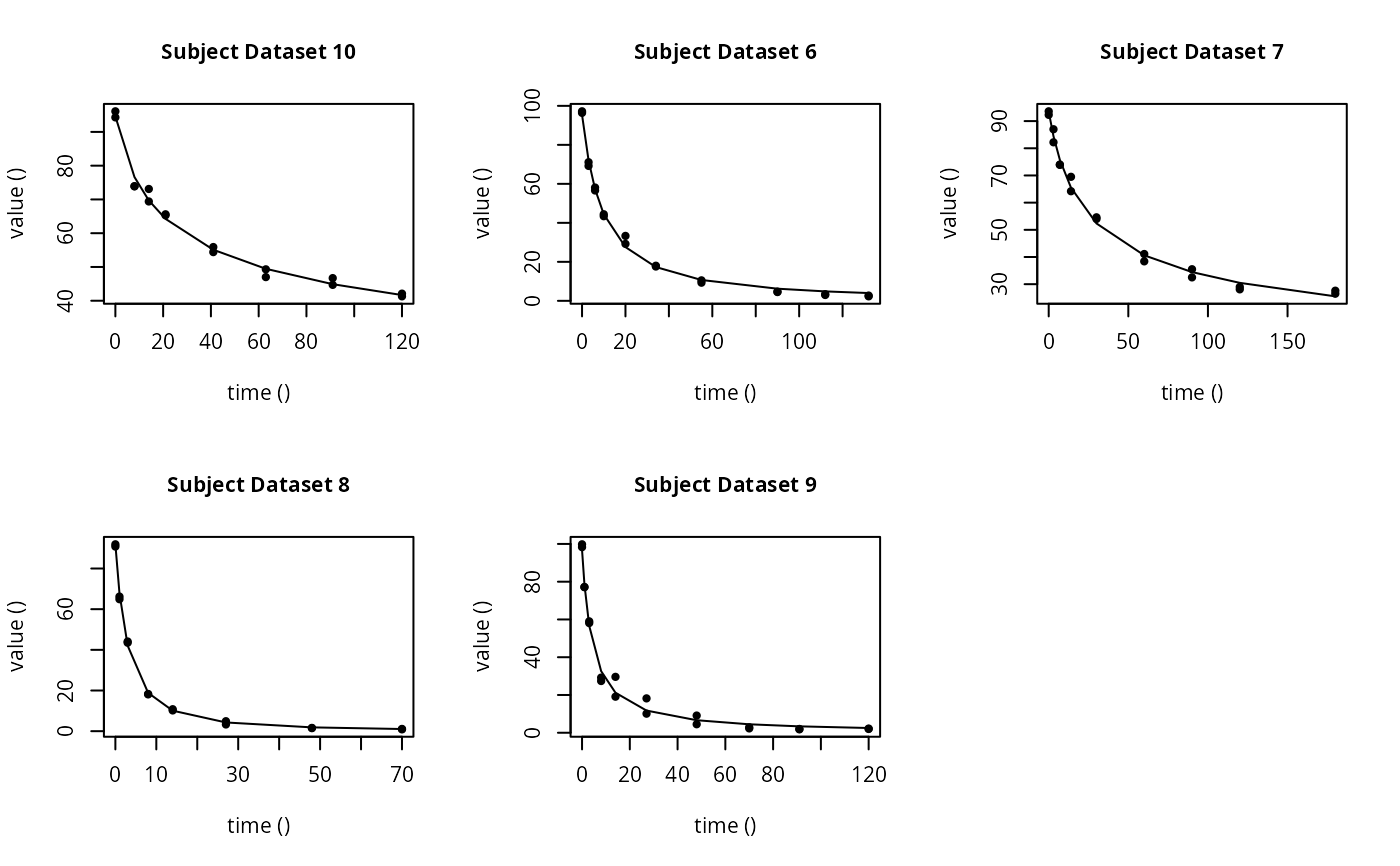
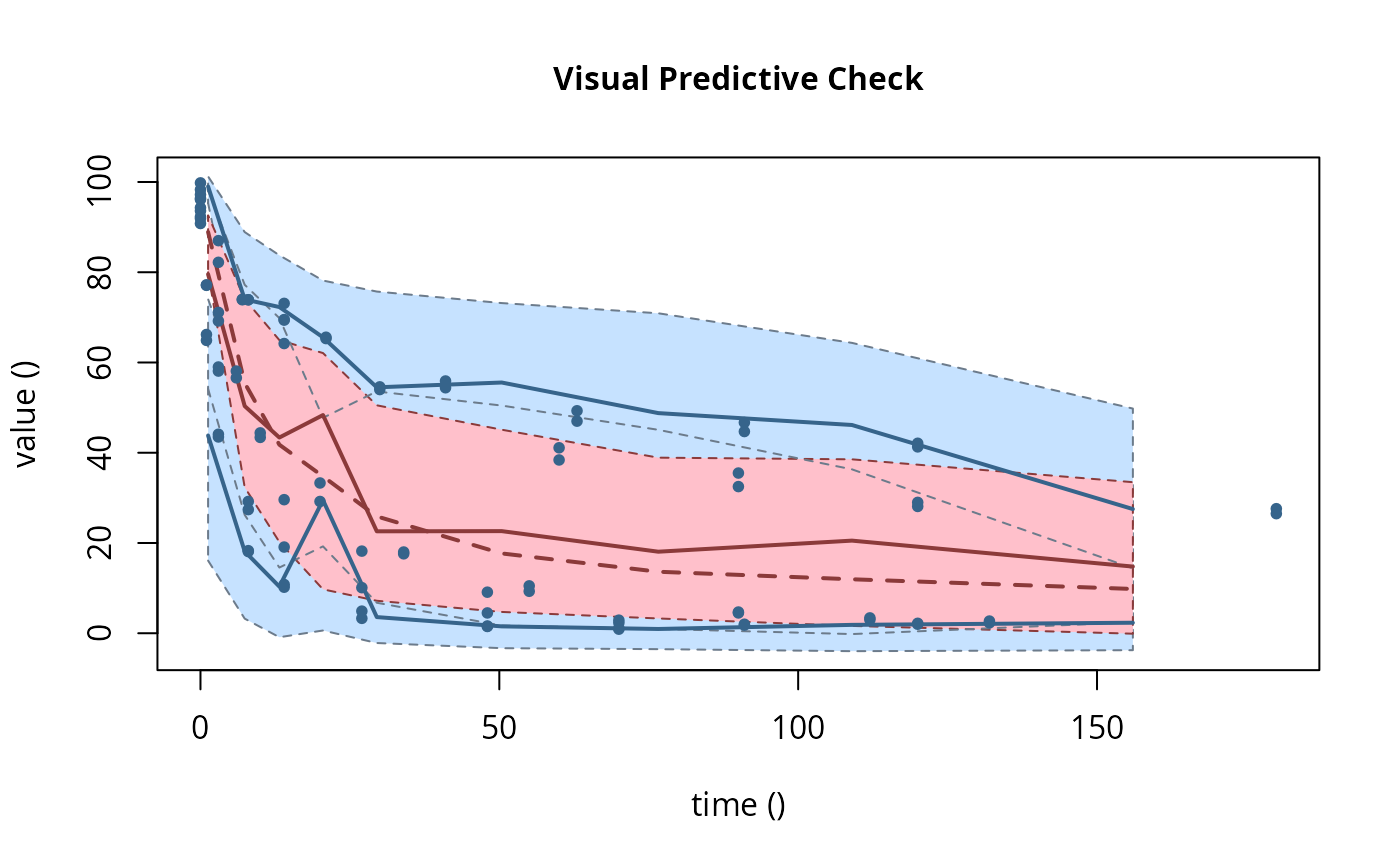
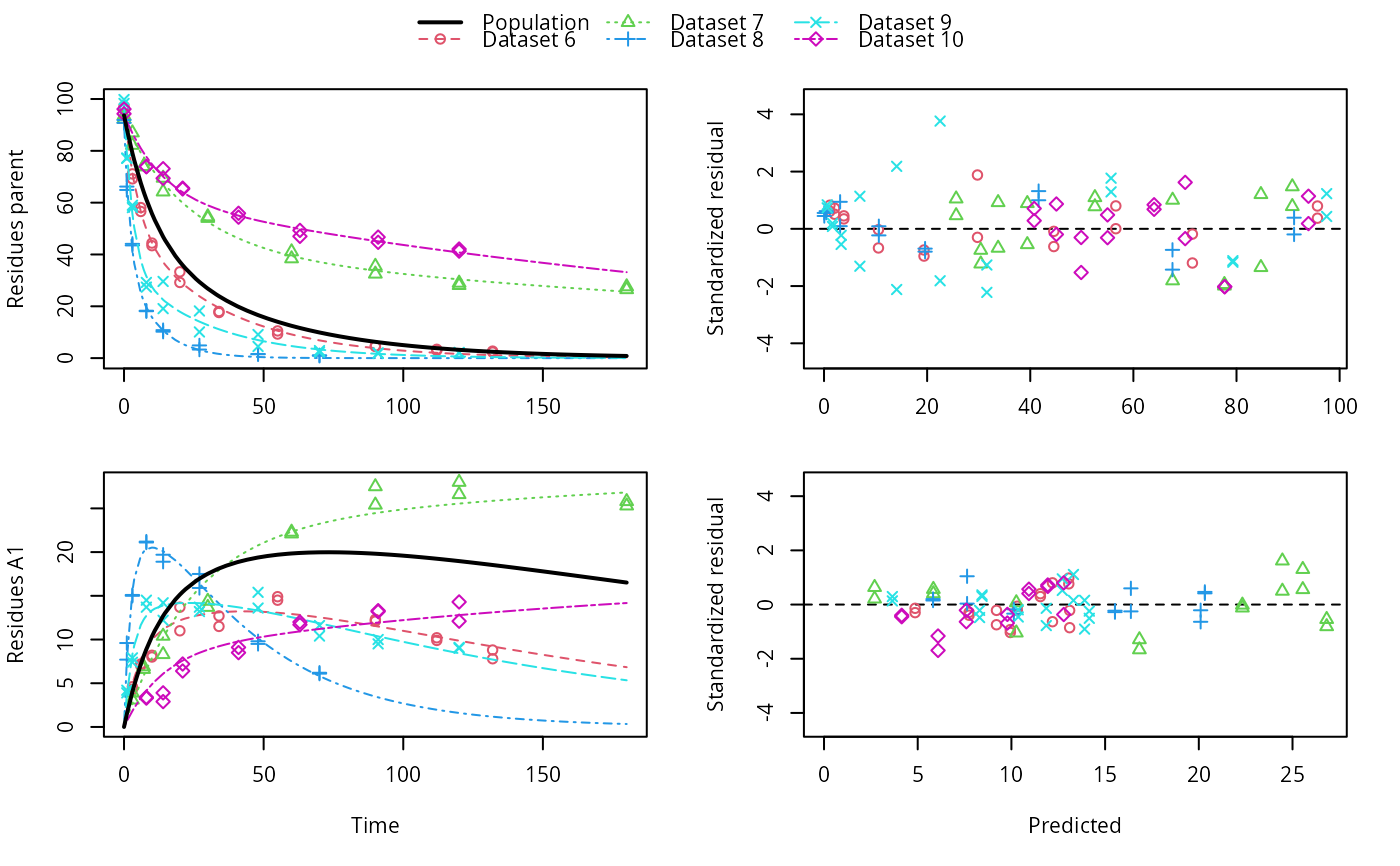
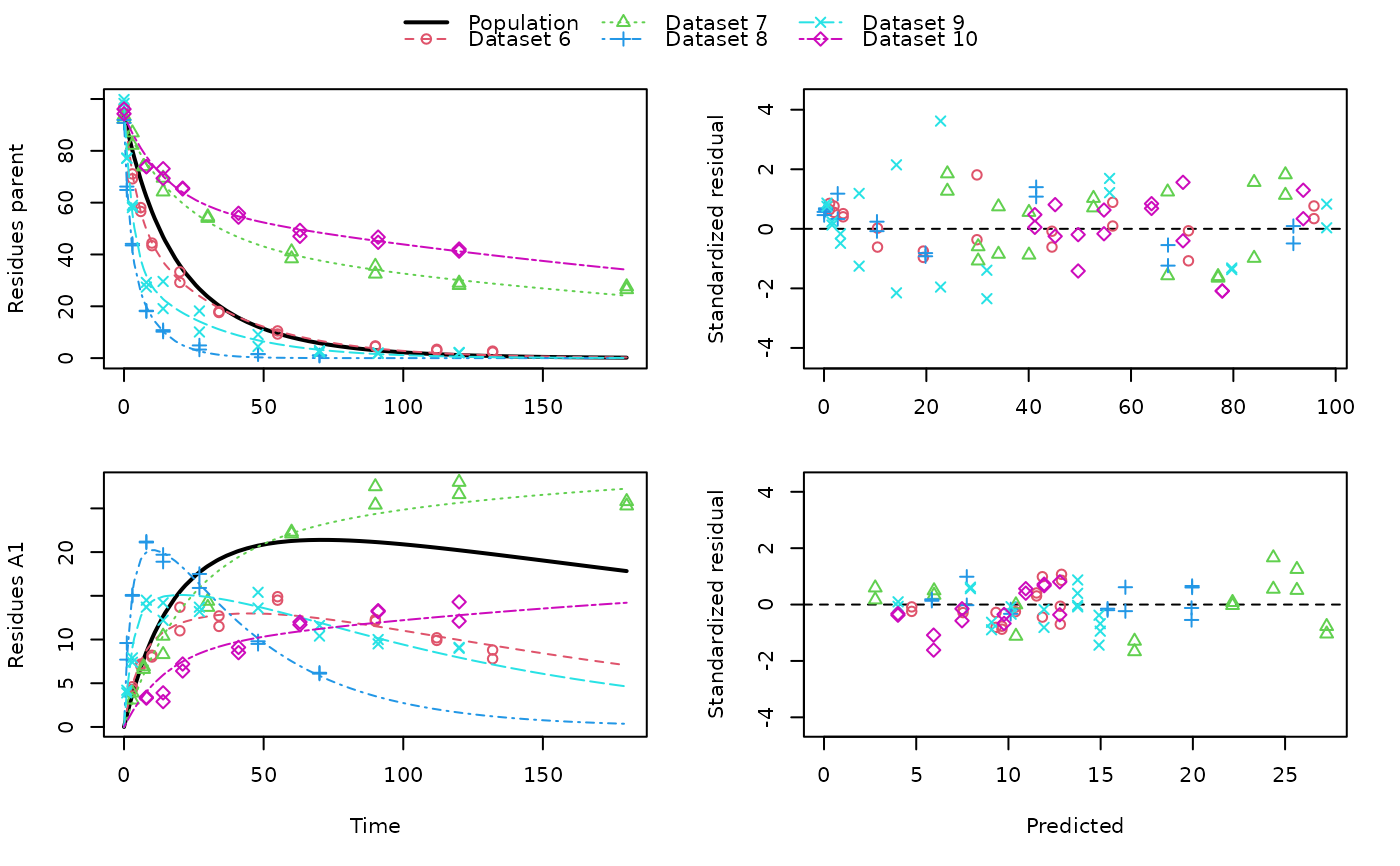
# \dontrun{ ds <- lapply(experimental_data_for_UBA_2019[6:10], function(x) subset(x$data[c("name", "time", "value")])) names(ds) <- paste("Dataset", 6:10) f_mmkin_parent_p0_fixed <- mmkin("FOMC", ds, cores = 1, state.ini = c(parent = 100), fixed_initials = "parent", quiet = TRUE) f_saem_p0_fixed <- saem(f_mmkin_parent_p0_fixed)#> Running main SAEM algorithm #> [1] "Wed Nov 11 19:47:58 2020" #> .... #> Minimisation finished #> [1] "Wed Nov 11 19:48:00 2020"f_mmkin_parent <- mmkin(c("SFO", "FOMC", "DFOP"), ds, quiet = TRUE) f_saem_sfo <- saem(f_mmkin_parent["SFO", ])#> Running main SAEM algorithm #> [1] "Wed Nov 11 19:48:01 2020" #> .... #> Minimisation finished #> [1] "Wed Nov 11 19:48:03 2020"f_saem_fomc <- saem(f_mmkin_parent["FOMC", ])#> Running main SAEM algorithm #> [1] "Wed Nov 11 19:48:03 2020" #> .... #> Minimisation finished #> [1] "Wed Nov 11 19:48:05 2020"f_saem_dfop <- saem(f_mmkin_parent["DFOP", ])#> Running main SAEM algorithm #> [1] "Wed Nov 11 19:48:06 2020" #> .... #> Minimisation finished #> [1] "Wed Nov 11 19:48:08 2020"# The returned saem.mmkin object contains an SaemixObject, therefore we can use # functions from saemix library(saemix)#> #>#> Likelihoods computed by importance sampling#> AIC BIC #> 1 624.2484 622.2956 #> 2 467.7096 464.9757 #> 3 495.4373 491.9222#> Plotting convergence plots#> Plotting individual fits#> Simulating data using nsim = 1000 simulated datasets #> Computing WRES and npde . #> Plotting npde#> --------------------------------------------- #> Distribution of npde: #> mean= -0.01528 (SE= 0.098 ) #> variance= 0.862 (SE= 0.13 ) #> skewness= 0.5016 #> kurtosis= 1.18 #> --------------------------------------------- #> #> Statistical tests #> Wilcoxon signed rank test : 0.679 #> Fisher variance test : 0.36 #> SW test of normality : 0.0855 . #> Global adjusted p-value : 0.257 #> --- #> Signif. codes: '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 #> ---------------------------------------------#> Performing simulations under the model. #> Plotting VPC #> Method used for VPC: binning by quantiles on X , dividing into the following intervals #> Interval Centered.On #> 1 (-1,3] 1.3 #> 2 (3,8] 7.4 #> 3 (8,14] 13.2 #> 4 (14,21] 20.5 #> 5 (21,37.7] 29.5 #> 6 (37.7,60] 50.4 #> 7 (60,90] 76.6 #> 8 (90,120] 109.0 #> 9 (120,180] 156.0f_mmkin_parent_tc <- update(f_mmkin_parent, error_model = "tc") f_saem_fomc_tc <- saem(f_mmkin_parent_tc["FOMC", ])#> Running main SAEM algorithm #> [1] "Wed Nov 11 19:48:11 2020" #> .... #> Minimisation finished #> [1] "Wed Nov 11 19:48:16 2020"#> Likelihoods computed by importance sampling#> AIC BIC #> 1 467.7096 464.9757 #> 2 469.5208 466.3963#>#>#># The following fit uses analytical solutions for SFO-SFO and DFOP-SFO, # and compiled ODEs for FOMC, both are fast f_mmkin <- mmkin(list( "SFO-SFO" = sfo_sfo, "FOMC-SFO" = fomc_sfo, "DFOP-SFO" = dfop_sfo), ds, quiet = TRUE) # These take about five seconds each on this system, as we use # analytical solutions written for saemix. When using the analytical # solutions written for mkin this took around four minutes f_saem_sfo_sfo <- saem(f_mmkin["SFO-SFO", ])#> Running main SAEM algorithm #> [1] "Wed Nov 11 19:48:18 2020" #> .... #> Minimisation finished #> [1] "Wed Nov 11 19:48:23 2020"f_saem_dfop_sfo <- saem(f_mmkin["DFOP-SFO", ])#> Running main SAEM algorithm #> [1] "Wed Nov 11 19:48:23 2020" #> .... #> Minimisation finished #> [1] "Wed Nov 11 19:48:32 2020"#> Kinetic nonlinear mixed-effects model fit by SAEM #> Structural model: #> d_parent/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 * #> time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time))) #> * parent #> d_A1/dt = + f_parent_to_A1 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g) #> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) * #> exp(-k2 * time))) * parent - k_A1 * A1 #> #> Data: #> 170 observations of 2 variable(s) grouped in 5 datasets #> #> Likelihood computed by importance sampling #> #> LL by is "-407.78 (df=13)" #> AIC BIC logLik #> 841.6 836.5 -407.8 #> #> Fitted parameters: #> estimate lower upper #> parent_0 93.76647 91.15312 96.3798 #> log_k_A1 -6.13235 -8.45788 -3.8068 #> f_parent_qlogis -0.97364 -1.36940 -0.5779 #> log_k1 -2.53176 -3.80372 -1.2598 #> log_k2 -3.58667 -5.29524 -1.8781 #> g_qlogis 0.01238 -1.07968 1.1044 #> Var.parent_0 7.61106 -3.34955 18.5717 #> Var.log_k_A1 4.64679 -2.73133 12.0249 #> Var.f_parent_qlogis 0.19693 -0.05498 0.4488 #> Var.log_k1 2.01717 -0.51980 4.5542 #> Var.log_k2 3.63412 -0.92964 8.1979 #> Var.g_qlogis 0.20045 -0.97425 1.3751 #> a.1 1.88335 1.66636 2.1004 #> SD.parent_0 2.75881 0.77234 4.7453 #> SD.log_k_A1 2.15564 0.44429 3.8670 #> SD.f_parent_qlogis 0.44377 0.15994 0.7276 #> SD.log_k1 1.42027 0.52714 2.3134 #> SD.log_k2 1.90634 0.70934 3.1033 #> SD.g_qlogis 0.44771 -0.86417 1.7596#> #> LL by is "-407.78 (df=13)"#> saemix version used for fitting: 3.1.9000 #> mkin version used for pre-fitting: 0.9.50.4 #> R version used for fitting: 4.0.3 #> Date of fit: Wed Nov 11 19:48:33 2020 #> Date of summary: Wed Nov 11 19:48:33 2020 #> #> Equations: #> d_parent/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 * #> time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time))) #> * parent #> d_A1/dt = + f_parent_to_A1 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g) #> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) * #> exp(-k2 * time))) * parent - k_A1 * A1 #> #> Data: #> 170 observations of 2 variable(s) grouped in 5 datasets #> #> Model predictions using solution type analytical #> #> Fitted in 9.564 s using 300, 100 iterations #> #> Variance model: Constant variance #> #> Mean of starting values for individual parameters: #> parent_0 log_k_A1 f_parent_qlogis log_k1 log_k2 #> 93.8101519 -9.7647455 -0.9711148 -1.8799371 -4.2708142 #> g_qlogis #> 0.1356441 #> #> Fixed degradation parameter values: #> None #> #> Results: #> #> Likelihood computed by importance sampling #> AIC BIC logLik #> 841.6 836.5 -407.8 #> #> Optimised, transformed parameters with symmetric confidence intervals: #> est. lower upper #> parent_0 93.76647 91.153 96.3798 #> log_k_A1 -6.13235 -8.458 -3.8068 #> f_parent_qlogis -0.97364 -1.369 -0.5779 #> log_k1 -2.53176 -3.804 -1.2598 #> log_k2 -3.58667 -5.295 -1.8781 #> g_qlogis 0.01238 -1.080 1.1044 #> #> Correlation: #> prnt_0 lg__A1 f_prn_ log_k1 log_k2 #> log_k_A1 -0.013 #> f_parent_qlogis -0.025 0.050 #> log_k1 0.030 0.000 -0.005 #> log_k2 0.010 0.005 -0.003 0.032 #> g_qlogis -0.063 -0.015 0.010 -0.167 -0.177 #> #> Random effects: #> est. lower upper #> SD.parent_0 2.7588 0.7723 4.7453 #> SD.log_k_A1 2.1556 0.4443 3.8670 #> SD.f_parent_qlogis 0.4438 0.1599 0.7276 #> SD.log_k1 1.4203 0.5271 2.3134 #> SD.log_k2 1.9063 0.7093 3.1033 #> SD.g_qlogis 0.4477 -0.8642 1.7596 #> #> Variance model: #> est. lower upper #> a.1 1.883 1.666 2.1 #> #> Backtransformed parameters with asymmetric confidence intervals: #> est. lower upper #> parent_0 93.766473 9.115e+01 96.37983 #> k_A1 0.002171 2.122e-04 0.02222 #> f_parent_to_A1 0.274156 2.027e-01 0.35942 #> k1 0.079519 2.229e-02 0.28371 #> k2 0.027691 5.015e-03 0.15288 #> g 0.503095 2.536e-01 0.75109 #> #> Resulting formation fractions: #> ff #> parent_A1 0.2742 #> parent_sink 0.7258 #> #> Estimated disappearance times: #> DT50 DT90 DT50back DT50_k1 DT50_k2 #> parent 14.11 59.53 17.92 8.717 25.03 #> A1 319.21 1060.38 NA NA NA# Using a single core, the following takes about 6 minutes as we do not have an # analytical solution. Using 10 cores it is slower instead of faster #f_saem_fomc <- saem(f_mmkin["FOMC-SFO", ], cores = 1) # }