Aerobic soil degradation data on dimethenamid and dimethenamid-P from the EU assessment in 2018
Source:R/dimethenamid_2018.R
dimethenamid_2018.RdThe datasets were extracted from the active substance evaluation dossier published by EFSA. Kinetic evaluations shown for these datasets are intended to illustrate and advance kinetic modelling. The fact that these data and some results are shown here does not imply a license to use them in the context of pesticide registrations, as the use of the data may be constrained by data protection regulations.
dimethenamid_2018Format
An mkindsg object grouping seven datasets with some meta information
Source
Rapporteur Member State Germany, Co-Rapporteur Member State Bulgaria (2018) Renewal Assessment Report Dimethenamid-P Volume 3 - B.8 Environmental fate and behaviour Rev. 2 - November 2017 https://open.efsa.europa.eu/study-inventory/EFSA-Q-2014-00716
Details
The R code used to create this data object is installed with this package in the 'dataset_generation' directory. In the code, page numbers are given for specific pieces of information in the comments.
Examples
print(dimethenamid_2018)
#> <mkindsg> holding 7 mkinds objects
#> Title $title: Aerobic soil degradation data on dimethenamid-P from the EU assessment in 2018
#> Occurrence of observed compounds $observed_n:
#> DMTAP M23 M27 M31 DMTA
#> 3 7 7 7 4
#> Time normalisation factors $f_time_norm:
#> [1] 1.0000000 0.9706477 1.2284784 1.2284784 0.6233856 0.7678922 0.6733938
#> Meta information $meta:
#> study usda_soil_type study_moisture_ref_type rel_moisture
#> Calke Unsworth 2014 Sandy loam pF2 1.00
#> Borstel Staudenmaier 2009 Sand pF1 0.50
#> Elliot 1 Wendt 1997 Clay loam pF2.5 0.75
#> Elliot 2 Wendt 1997 Clay loam pF2.5 0.75
#> Flaach König 1996 Sandy clay loam pF1 0.40
#> BBA 2.2 König 1995 Loamy sand pF1 0.40
#> BBA 2.3 König 1995 Sandy loam pF1 0.40
#> study_ref_moisture temperature
#> Calke NA 20
#> Borstel 23.00 20
#> Elliot 1 33.37 23
#> Elliot 2 33.37 23
#> Flaach NA 20
#> BBA 2.2 NA 20
#> BBA 2.3 NA 20
dmta_ds <- lapply(1:7, function(i) {
ds_i <- dimethenamid_2018$ds[[i]]$data
ds_i[ds_i$name == "DMTAP", "name"] <- "DMTA"
ds_i$time <- ds_i$time * dimethenamid_2018$f_time_norm[i]
ds_i
})
names(dmta_ds) <- sapply(dimethenamid_2018$ds, function(ds) ds$title)
dmta_ds[["Elliot"]] <- rbind(dmta_ds[["Elliot 1"]], dmta_ds[["Elliot 2"]])
dmta_ds[["Elliot 1"]] <- NULL
dmta_ds[["Elliot 2"]] <- NULL
# \dontrun{
dfop_sfo3_plus <- mkinmod(
DMTA = mkinsub("DFOP", c("M23", "M27", "M31")),
M23 = mkinsub("SFO"),
M27 = mkinsub("SFO"),
M31 = mkinsub("SFO", "M27", sink = FALSE),
quiet = TRUE
)
f_dmta_mkin_tc <- mmkin(
list("DFOP-SFO3+" = dfop_sfo3_plus),
dmta_ds, quiet = TRUE, error_model = "tc")
nlmixr_model(f_dmta_mkin_tc)
#> With est = 'saem', a different error model is required for each observed variableChanging the error model to 'obs_tc' (Two-component error for each observed variable)
#> function ()
#> {
#> ini({
#> DMTA_0 = 99
#> eta.DMTA_0 ~ 2.3
#> log_k_M23 = -3.9
#> eta.log_k_M23 ~ 0.55
#> log_k_M27 = -4.3
#> eta.log_k_M27 ~ 0.86
#> log_k_M31 = -4.2
#> eta.log_k_M31 ~ 0.75
#> log_k1 = -2.2
#> eta.log_k1 ~ 0.9
#> log_k2 = -3.8
#> eta.log_k2 ~ 1.6
#> g_qlogis = 0.44
#> eta.g_qlogis ~ 3.1
#> f_DMTA_tffm0_1_qlogis = -2.1
#> eta.f_DMTA_tffm0_1_qlogis ~ 0.3
#> f_DMTA_tffm0_2_qlogis = -2.2
#> eta.f_DMTA_tffm0_2_qlogis ~ 0.3
#> f_DMTA_tffm0_3_qlogis = -2.1
#> eta.f_DMTA_tffm0_3_qlogis ~ 0.3
#> sigma_low_DMTA = 0.7
#> rsd_high_DMTA = 0.026
#> sigma_low_M23 = 0.7
#> rsd_high_M23 = 0.026
#> sigma_low_M27 = 0.7
#> rsd_high_M27 = 0.026
#> sigma_low_M31 = 0.7
#> rsd_high_M31 = 0.026
#> })
#> model({
#> DMTA_0_model = DMTA_0 + eta.DMTA_0
#> DMTA(0) = DMTA_0_model
#> k_M23 = exp(log_k_M23 + eta.log_k_M23)
#> k_M27 = exp(log_k_M27 + eta.log_k_M27)
#> k_M31 = exp(log_k_M31 + eta.log_k_M31)
#> k1 = exp(log_k1 + eta.log_k1)
#> k2 = exp(log_k2 + eta.log_k2)
#> g = expit(g_qlogis + eta.g_qlogis)
#> f_DMTA_tffm0_1 = expit(f_DMTA_tffm0_1_qlogis + eta.f_DMTA_tffm0_1_qlogis)
#> f_DMTA_tffm0_2 = expit(f_DMTA_tffm0_2_qlogis + eta.f_DMTA_tffm0_2_qlogis)
#> f_DMTA_tffm0_3 = expit(f_DMTA_tffm0_3_qlogis + eta.f_DMTA_tffm0_3_qlogis)
#> f_DMTA_to_M23 = f_DMTA_tffm0_1
#> f_DMTA_to_M27 = f_DMTA_tffm0_2 * (1 - f_DMTA_tffm0_1)
#> f_DMTA_to_M31 = f_DMTA_tffm0_3 * (1 - f_DMTA_tffm0_2) *
#> (1 - f_DMTA_tffm0_1)
#> d/dt(DMTA) = -((k1 * g * exp(-k1 * time) + k2 * (1 -
#> g) * exp(-k2 * time))/(g * exp(-k1 * time) + (1 -
#> g) * exp(-k2 * time))) * DMTA
#> d/dt(M23) = +f_DMTA_to_M23 * ((k1 * g * exp(-k1 * time) +
#> k2 * (1 - g) * exp(-k2 * time))/(g * exp(-k1 * time) +
#> (1 - g) * exp(-k2 * time))) * DMTA - k_M23 * M23
#> d/dt(M27) = +f_DMTA_to_M27 * ((k1 * g * exp(-k1 * time) +
#> k2 * (1 - g) * exp(-k2 * time))/(g * exp(-k1 * time) +
#> (1 - g) * exp(-k2 * time))) * DMTA - k_M27 * M27 +
#> k_M31 * M31
#> d/dt(M31) = +f_DMTA_to_M31 * ((k1 * g * exp(-k1 * time) +
#> k2 * (1 - g) * exp(-k2 * time))/(g * exp(-k1 * time) +
#> (1 - g) * exp(-k2 * time))) * DMTA - k_M31 * M31
#> DMTA ~ add(sigma_low_DMTA) + prop(rsd_high_DMTA)
#> M23 ~ add(sigma_low_M23) + prop(rsd_high_M23)
#> M27 ~ add(sigma_low_M27) + prop(rsd_high_M27)
#> M31 ~ add(sigma_low_M31) + prop(rsd_high_M31)
#> })
#> }
#> <environment: 0x555560091f40>
# The focei fit takes about four minutes on my system
system.time(
f_dmta_nlmixr_focei <- nlmixr(f_dmta_mkin_tc, est = "focei",
control = nlmixr::foceiControl(print = 500))
)
#> ℹ parameter labels from comments are typically ignored in non-interactive mode
#> ℹ Need to run with the source intact to parse comments
#> → creating full model...
#> → pruning branches (`if`/`else`)...
#> ✔ done
#> → loading into symengine environment...
#> ✔ done
#> → creating full model...
#> → pruning branches (`if`/`else`)...
#> ✔ done
#> → loading into symengine environment...
#> ✔ done
#> → calculate jacobian
#> [====|====|====|====|====|====|====|====|====|====] 0:00:02
#>
#> → calculate sensitivities
#> [====|====|====|====|====|====|====|====|====|====] 0:00:04
#>
#> → calculate ∂(f)/∂(η)
#> [====|====|====|====|====|====|====|====|====|====] 0:00:01
#>
#> → calculate ∂(R²)/∂(η)
#> [====|====|====|====|====|====|====|====|====|====] 0:00:08
#>
#> → finding duplicate expressions in inner model...
#> [====|====|====|====|====|====|====|====|====|====] 0:00:07
#>
#> → optimizing duplicate expressions in inner model...
#> [====|====|====|====|====|====|====|====|====|====] 0:00:07
#>
#> → finding duplicate expressions in EBE model...
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> → optimizing duplicate expressions in EBE model...
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> → compiling inner model...
#>
#> ✔ done
#> → finding duplicate expressions in FD model...
#>
#> → optimizing duplicate expressions in FD model...
#>
#> → compiling EBE model...
#>
#> ✔ done
#> → compiling events FD model...
#>
#> ✔ done
#> Needed Covariates:
#> [1] "CMT"
#> RxODE 1.1.4 using 8 threads (see ?getRxThreads)
#> no cache: create with `rxCreateCache()`
#> Key: U: Unscaled Parameters; X: Back-transformed parameters; G: Gill difference gradient approximation
#> F: Forward difference gradient approximation
#> C: Central difference gradient approximation
#> M: Mixed forward and central difference gradient approximation
#> Unscaled parameters for Omegas=chol(solve(omega));
#> Diagonals are transformed, as specified by foceiControl(diagXform=)
#> |-----+---------------+-----------+-----------+-----------+-----------|
#> | #| Objective Fun | DMTA_0 | log_k_M23 | log_k_M27 | log_k_M31 |
#> |.....................| log_k1 | log_k2 | g_qlogis |f_DMTA_tffm0_1_qlogis |
#> |.....................|f_DMTA_tffm0_2_qlogis |f_DMTA_tffm0_3_qlogis | sigma_low | rsd_high |
#> |.....................| o1 | o2 | o3 | o4 |
#> |.....................| o5 | o6 | o7 | o8 |
#> |.....................| o9 | o10 |...........|...........|
#> calculating covariance matrix
#> done
#> Calculating residuals/tables
#> done
#> Warning: initial ETAs were nudged; (can control by foceiControl(etaNudge=., etaNudge2=))
#> Warning: ETAs were reset to zero during optimization; (Can control by foceiControl(resetEtaP=.))
#> Warning: last objective function was not at minimum, possible problems in optimization
#> Warning: S matrix non-positive definite
#> Warning: using R matrix to calculate covariance
#> Warning: gradient problems with initial estimate and covariance; see $scaleInfo
#> user system elapsed
#> 553.721 10.570 564.258
summary(f_dmta_nlmixr_focei)
#> nlmixr version used for fitting: 2.0.6
#> mkin version used for pre-fitting: 1.1.0
#> R version used for fitting: 4.1.2
#> Date of fit: Wed Mar 2 13:27:22 2022
#> Date of summary: Wed Mar 2 13:27:22 2022
#>
#> Equations:
#> d_DMTA/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 *
#> time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time)))
#> * DMTA
#> d_M23/dt = + f_DMTA_to_M23 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g)
#> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
#> exp(-k2 * time))) * DMTA - k_M23 * M23
#> d_M27/dt = + f_DMTA_to_M27 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g)
#> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
#> exp(-k2 * time))) * DMTA - k_M27 * M27 + k_M31 * M31
#> d_M31/dt = + f_DMTA_to_M31 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g)
#> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
#> exp(-k2 * time))) * DMTA - k_M31 * M31
#>
#> Data:
#> 563 observations of 4 variable(s) grouped in 6 datasets
#>
#> Degradation model predictions using RxODE
#>
#> Fitted in 564.08 s
#>
#> Variance model: Two-component variance function
#>
#> Mean of starting values for individual parameters:
#> DMTA_0 log_k_M23 log_k_M27 log_k_M31 f_DMTA_ilr_1 f_DMTA_ilr_2
#> 98.7132 -3.9216 -4.3306 -4.2442 0.1376 0.1388
#> f_DMTA_ilr_3 log_k1 log_k2 g_qlogis
#> -1.7554 -2.2352 -3.7758 0.4363
#>
#> Mean of starting values for error model parameters:
#> sigma_low rsd_high
#> 0.70012 0.02577
#>
#> Fixed degradation parameter values:
#> None
#>
#> Results:
#>
#> Likelihood calculated by focei
#> AIC BIC logLik
#> 1857 1952 -906.5
#>
#> Optimised parameters:
#> est. lower upper
#> DMTA_0 98.0116 95.243 100.780
#> log_k_M23 -4.0184 -5.213 -2.824
#> log_k_M27 -4.2033 -5.013 -3.394
#> log_k_M31 -4.1728 -4.999 -3.347
#> log_k1 -2.4831 -3.398 -1.568
#> log_k2 -3.8423 -5.450 -2.235
#> g_qlogis 0.4682 -2.188 3.124
#> f_DMTA_tffm0_1_qlogis -2.0823 -2.591 -1.574
#> f_DMTA_tffm0_2_qlogis -2.1265 -2.686 -1.567
#> f_DMTA_tffm0_3_qlogis -2.0795 -2.735 -1.424
#>
#> Correlation:
#> DMTA_0 lg__M23 lg__M27 lg__M31 log_k1 log_k2 g_qlogs
#> log_k_M23 -0.0154
#> log_k_M27 -0.0164 0.0031
#> log_k_M31 -0.0131 0.0018 0.0541
#> log_k1 -0.0306 0.0045 0.0019 0.0011
#> log_k2 0.0527 -0.0043 -0.0037 -0.0003 0.0375
#> g_qlogis -0.1005 0.0076 0.0074 0.0013 0.0910 0.1151
#> f_DMTA_tffm0_1_qlogis -0.0308 0.0362 0.0024 0.0021 0.0058 -0.0070 0.0145
#> f_DMTA_tffm0_2_qlogis -0.0309 0.0062 0.0353 -0.0229 0.0047 -0.0082 0.0146
#> f_DMTA_tffm0_3_qlogis -0.0308 0.0061 0.0419 0.0547 0.0033 -0.0055 0.0104
#> f_DMTA_0_1 f_DMTA_0_2
#> log_k_M23
#> log_k_M27
#> log_k_M31
#> log_k1
#> log_k2
#> g_qlogis
#> f_DMTA_tffm0_1_qlogis
#> f_DMTA_tffm0_2_qlogis 0.0118
#> f_DMTA_tffm0_3_qlogis 0.0086 -0.0057
#>
#> Random effects (omega):
#> eta.DMTA_0 eta.log_k_M23 eta.log_k_M27 eta.log_k_M31
#> eta.DMTA_0 4.224 0.000 0.0000 0.0000
#> eta.log_k_M23 0.000 1.041 0.0000 0.0000
#> eta.log_k_M27 0.000 0.000 0.4609 0.0000
#> eta.log_k_M31 0.000 0.000 0.0000 0.4728
#> eta.log_k1 0.000 0.000 0.0000 0.0000
#> eta.log_k2 0.000 0.000 0.0000 0.0000
#> eta.g_qlogis 0.000 0.000 0.0000 0.0000
#> eta.f_DMTA_tffm0_1_qlogis 0.000 0.000 0.0000 0.0000
#> eta.f_DMTA_tffm0_2_qlogis 0.000 0.000 0.0000 0.0000
#> eta.f_DMTA_tffm0_3_qlogis 0.000 0.000 0.0000 0.0000
#> eta.log_k1 eta.log_k2 eta.g_qlogis
#> eta.DMTA_0 0.000 0.000 0.00
#> eta.log_k_M23 0.000 0.000 0.00
#> eta.log_k_M27 0.000 0.000 0.00
#> eta.log_k_M31 0.000 0.000 0.00
#> eta.log_k1 0.635 0.000 0.00
#> eta.log_k2 0.000 1.662 0.00
#> eta.g_qlogis 0.000 0.000 4.36
#> eta.f_DMTA_tffm0_1_qlogis 0.000 0.000 0.00
#> eta.f_DMTA_tffm0_2_qlogis 0.000 0.000 0.00
#> eta.f_DMTA_tffm0_3_qlogis 0.000 0.000 0.00
#> eta.f_DMTA_tffm0_1_qlogis eta.f_DMTA_tffm0_2_qlogis
#> eta.DMTA_0 0.0000 0.0000
#> eta.log_k_M23 0.0000 0.0000
#> eta.log_k_M27 0.0000 0.0000
#> eta.log_k_M31 0.0000 0.0000
#> eta.log_k1 0.0000 0.0000
#> eta.log_k2 0.0000 0.0000
#> eta.g_qlogis 0.0000 0.0000
#> eta.f_DMTA_tffm0_1_qlogis 0.1909 0.0000
#> eta.f_DMTA_tffm0_2_qlogis 0.0000 0.2232
#> eta.f_DMTA_tffm0_3_qlogis 0.0000 0.0000
#> eta.f_DMTA_tffm0_3_qlogis
#> eta.DMTA_0 0.0000
#> eta.log_k_M23 0.0000
#> eta.log_k_M27 0.0000
#> eta.log_k_M31 0.0000
#> eta.log_k1 0.0000
#> eta.log_k2 0.0000
#> eta.g_qlogis 0.0000
#> eta.f_DMTA_tffm0_1_qlogis 0.0000
#> eta.f_DMTA_tffm0_2_qlogis 0.0000
#> eta.f_DMTA_tffm0_3_qlogis 0.3149
#>
#> Variance model:
#> sigma_low rsd_high
#> 0.82408 0.03045
#>
#> Backtransformed parameters:
#> est. lower upper
#> DMTA_0 98.01163 95.243379 100.77988
#> k_M23 0.01798 0.005443 0.05940
#> k_M27 0.01495 0.006652 0.03358
#> k_M31 0.01541 0.006746 0.03520
#> f_DMTA_to_M23 0.11083 NA NA
#> f_DMTA_to_M27 0.09474 NA NA
#> f_DMTA_to_M31 0.08827 NA NA
#> k1 0.08348 0.033429 0.20848
#> k2 0.02144 0.004296 0.10704
#> g 0.61496 0.100857 0.95788
#>
#> Resulting formation fractions:
#> ff
#> DMTA_M23 0.11083
#> DMTA_M27 0.09474
#> DMTA_M31 0.08827
#> DMTA_sink 0.70616
#>
#> Estimated disappearance times:
#> DT50 DT90 DT50back DT50_k1 DT50_k2
#> DMTA 12.96 64.24 19.34 8.303 32.32
#> M23 38.55 128.06 NA NA NA
#> M27 46.38 154.06 NA NA NA
#> M31 44.98 149.43 NA NA NA
plot(f_dmta_nlmixr_focei)
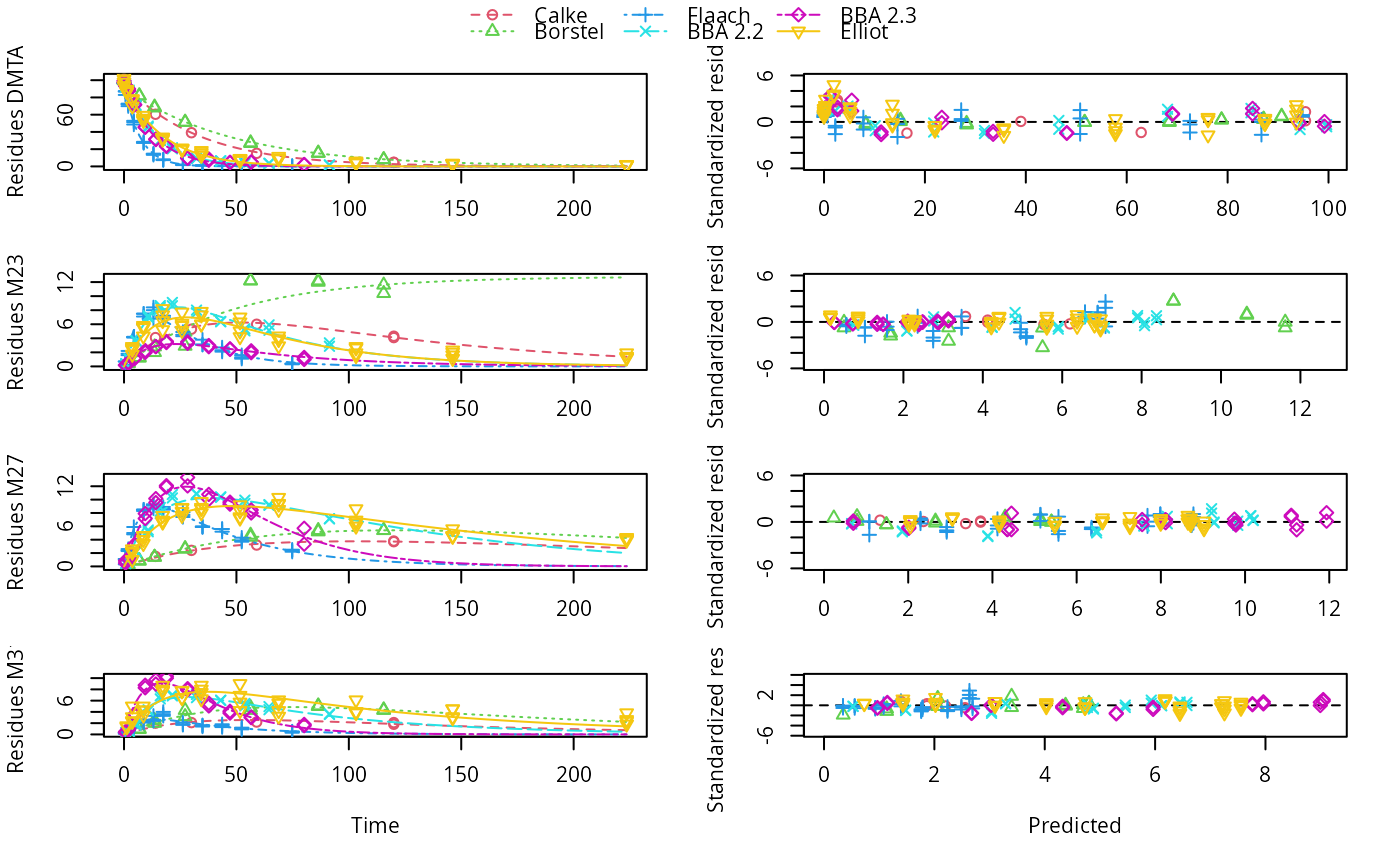 # Using saemix takes about 18 minutes
system.time(
f_dmta_saemix <- saem(f_dmta_mkin_tc, test_log_parms = TRUE)
)
#> DINTDY- T (=R1) illegal
#> In above message, R1 = 115.507
#>
#> T not in interval TCUR - HU (= R1) to TCUR (=R2)
#> In above message, R1 = 112.133, R2 = 113.577
#>
#> DLSODA- At T (=R1), too much accuracy requested
#> for precision of machine.. See TOLSF (=R2)
#> In above message, R1 = 55.3899, R2 = nan
#>
#> Error in out[available, var]: (subscript) logical subscript too long
#> Timing stopped at: 12.58 0 12.58
#> Timing stopped at: 12.99 0.008 13
# nlmixr with est = "saem" is pretty fast with default iteration numbers, most
# of the time (about 2.5 minutes) is spent for calculating the log likelihood at the end
# The likelihood calculated for the nlmixr fit is much lower than that found by saemix
# Also, the trace plot and the plot of the individual predictions is not
# convincing for the parent. It seems we are fitting an overparameterised
# model, so the result we get strongly depends on starting parameters and control settings.
system.time(
f_dmta_nlmixr_saem <- nlmixr(f_dmta_mkin_tc, est = "saem",
control = nlmixr::saemControl(print = 500, logLik = TRUE, nmc = 9))
)
#> With est = 'saem', a different error model is required for each observed variableChanging the error model to 'obs_tc' (Two-component error for each observed variable)
#> ℹ parameter labels from comments are typically ignored in non-interactive mode
#> ℹ Need to run with the source intact to parse comments
#>
#> → generate SAEM model
#> ✔ done
#> 1: 98.7179 -3.4492 -3.2592 -3.6952 -2.1629 -2.7824 0.8990 -2.8080 -2.7380 -2.8041 2.7789 0.6848 0.8170 0.7125 0.8550 1.5200 2.9882 0.3073 0.2850 0.2877 4.0480 0.4153 4.5214 0.3775 4.4419 0.4181 3.7069 0.5935
#> 500: 97.8519 -4.3891 -4.0888 -4.1247 -2.9246 -4.2755 2.6294 -2.1212 -2.1380 -2.0739 3.1293 1.2665 0.2763 0.3429 0.5743 1.5561 4.4991 0.1499 0.1551 0.3103 0.9514 0.0341 0.4846 0.1068 0.6597 0.0767 0.7836 0.0360
#> Calculating covariance matrix
#>
#> Calculating -2LL by Gaussian quadrature (nnodes=3,nsd=1.6)
#>
#> → creating full model...
#> → pruning branches (`if`/`else`)...
#> ✔ done
#> → loading into symengine environment...
#> ✔ done
#> → compiling EBE model...
#>
#> ✔ done
#> Needed Covariates:
#> [1] "CMT"
#> Calculating residuals/tables
#> done
#> user system elapsed
#> 785.825 3.841 153.598
traceplot(f_dmta_nlmixr_saem$nm)
#> Error in traceplot(f_dmta_nlmixr_saem$nm): could not find function "traceplot"
summary(f_dmta_nlmixr_saem)
#> nlmixr version used for fitting: 2.0.6
#> mkin version used for pre-fitting: 1.1.0
#> R version used for fitting: 4.1.2
#> Date of fit: Wed Mar 2 13:30:09 2022
#> Date of summary: Wed Mar 2 13:30:09 2022
#>
#> Equations:
#> d_DMTA/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 *
#> time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time)))
#> * DMTA
#> d_M23/dt = + f_DMTA_to_M23 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g)
#> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
#> exp(-k2 * time))) * DMTA - k_M23 * M23
#> d_M27/dt = + f_DMTA_to_M27 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g)
#> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
#> exp(-k2 * time))) * DMTA - k_M27 * M27 + k_M31 * M31
#> d_M31/dt = + f_DMTA_to_M31 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g)
#> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
#> exp(-k2 * time))) * DMTA - k_M31 * M31
#>
#> Data:
#> 563 observations of 4 variable(s) grouped in 6 datasets
#>
#> Degradation model predictions using RxODE
#>
#> Fitted in 153.313 s
#>
#> Variance model: Two-component variance function
#>
#> Mean of starting values for individual parameters:
#> DMTA_0 log_k_M23 log_k_M27 log_k_M31 f_DMTA_ilr_1 f_DMTA_ilr_2
#> 98.7132 -3.9216 -4.3306 -4.2442 0.1376 0.1388
#> f_DMTA_ilr_3 log_k1 log_k2 g_qlogis
#> -1.7554 -2.2352 -3.7758 0.4363
#>
#> Mean of starting values for error model parameters:
#> sigma_low_DMTA rsd_high_DMTA sigma_low_M23 rsd_high_M23 sigma_low_M27
#> 0.70012 0.02577 0.70012 0.02577 0.70012
#> rsd_high_M27 sigma_low_M31 rsd_high_M31
#> 0.02577 0.70012 0.02577
#>
#> Fixed degradation parameter values:
#> None
#>
#> Results:
#>
#> Likelihood calculated by focei
#> AIC BIC logLik
#> 1966 2088 -955.2
#>
#> Optimised parameters:
#> est. lower upper
#> DMTA_0 97.852 95.86386 99.840
#> log_k_M23 -4.389 -5.35084 -3.427
#> log_k_M27 -4.089 -4.54432 -3.633
#> log_k_M31 -4.125 -4.63280 -3.617
#> log_k1 -2.925 -3.54158 -2.308
#> log_k2 -4.275 -5.81760 -2.733
#> g_qlogis 2.629 -0.01785 5.277
#> f_DMTA_tffm0_1_qlogis -2.121 -2.44462 -1.798
#> f_DMTA_tffm0_2_qlogis -2.138 -2.47804 -1.798
#> f_DMTA_tffm0_3_qlogis -2.074 -2.53581 -1.612
#>
#> Correlation:
#> DMTA_0 lg__M23 lg__M27 lg__M31 log_k1 log_k2 g_qlogs
#> log_k_M23 -0.0164
#> log_k_M27 -0.0267 0.0028
#> log_k_M31 -0.0179 0.0023 0.0755
#> log_k1 0.0385 -0.0034 -0.0054 -0.0029
#> log_k2 0.0381 0.0115 0.0087 0.0093 0.0786
#> g_qlogis -0.0656 0.0021 0.0051 0.0001 -0.1177 -0.4389
#> f_DMTA_tffm0_1_qlogis -0.0604 0.0554 0.0054 0.0039 -0.0082 -0.0022 0.0119
#> f_DMTA_tffm0_2_qlogis -0.0601 0.0091 0.0577 -0.0350 -0.0081 -0.0057 0.0137
#> f_DMTA_tffm0_3_qlogis -0.0515 0.0083 0.0569 0.0729 -0.0059 0.0005 0.0073
#> f_DMTA_0_1 f_DMTA_0_2
#> log_k_M23
#> log_k_M27
#> log_k_M31
#> log_k1
#> log_k2
#> g_qlogis
#> f_DMTA_tffm0_1_qlogis
#> f_DMTA_tffm0_2_qlogis 0.0167
#> f_DMTA_tffm0_3_qlogis 0.0145 -0.0060
#>
#> Random effects (omega):
#> eta.DMTA_0 eta.log_k_M23 eta.log_k_M27 eta.log_k_M31
#> eta.DMTA_0 3.129 0.000 0.0000 0.0000
#> eta.log_k_M23 0.000 1.266 0.0000 0.0000
#> eta.log_k_M27 0.000 0.000 0.2763 0.0000
#> eta.log_k_M31 0.000 0.000 0.0000 0.3429
#> eta.log_k1 0.000 0.000 0.0000 0.0000
#> eta.log_k2 0.000 0.000 0.0000 0.0000
#> eta.g_qlogis 0.000 0.000 0.0000 0.0000
#> eta.f_DMTA_tffm0_1_qlogis 0.000 0.000 0.0000 0.0000
#> eta.f_DMTA_tffm0_2_qlogis 0.000 0.000 0.0000 0.0000
#> eta.f_DMTA_tffm0_3_qlogis 0.000 0.000 0.0000 0.0000
#> eta.log_k1 eta.log_k2 eta.g_qlogis
#> eta.DMTA_0 0.0000 0.000 0.000
#> eta.log_k_M23 0.0000 0.000 0.000
#> eta.log_k_M27 0.0000 0.000 0.000
#> eta.log_k_M31 0.0000 0.000 0.000
#> eta.log_k1 0.5743 0.000 0.000
#> eta.log_k2 0.0000 1.556 0.000
#> eta.g_qlogis 0.0000 0.000 4.499
#> eta.f_DMTA_tffm0_1_qlogis 0.0000 0.000 0.000
#> eta.f_DMTA_tffm0_2_qlogis 0.0000 0.000 0.000
#> eta.f_DMTA_tffm0_3_qlogis 0.0000 0.000 0.000
#> eta.f_DMTA_tffm0_1_qlogis eta.f_DMTA_tffm0_2_qlogis
#> eta.DMTA_0 0.0000 0.0000
#> eta.log_k_M23 0.0000 0.0000
#> eta.log_k_M27 0.0000 0.0000
#> eta.log_k_M31 0.0000 0.0000
#> eta.log_k1 0.0000 0.0000
#> eta.log_k2 0.0000 0.0000
#> eta.g_qlogis 0.0000 0.0000
#> eta.f_DMTA_tffm0_1_qlogis 0.1499 0.0000
#> eta.f_DMTA_tffm0_2_qlogis 0.0000 0.1551
#> eta.f_DMTA_tffm0_3_qlogis 0.0000 0.0000
#> eta.f_DMTA_tffm0_3_qlogis
#> eta.DMTA_0 0.0000
#> eta.log_k_M23 0.0000
#> eta.log_k_M27 0.0000
#> eta.log_k_M31 0.0000
#> eta.log_k1 0.0000
#> eta.log_k2 0.0000
#> eta.g_qlogis 0.0000
#> eta.f_DMTA_tffm0_1_qlogis 0.0000
#> eta.f_DMTA_tffm0_2_qlogis 0.0000
#> eta.f_DMTA_tffm0_3_qlogis 0.3103
#>
#> Variance model:
#> sigma_low_DMTA rsd_high_DMTA sigma_low_M23 rsd_high_M23 sigma_low_M27
#> 0.95135 0.03412 0.48455 0.10682 0.65969
#> rsd_high_M27 sigma_low_M31 rsd_high_M31
#> 0.07670 0.78365 0.03598
#>
#> Backtransformed parameters:
#> est. lower upper
#> DMTA_0 97.85189 95.863863 99.83992
#> k_M23 0.01241 0.004744 0.03247
#> k_M27 0.01676 0.010627 0.02643
#> k_M31 0.01617 0.009727 0.02687
#> f_DMTA_to_M23 0.10705 NA NA
#> f_DMTA_to_M27 0.09417 NA NA
#> f_DMTA_to_M31 0.08919 NA NA
#> k1 0.05369 0.028968 0.09950
#> k2 0.01391 0.002975 0.06500
#> g 0.93273 0.495538 0.99492
#>
#> Resulting formation fractions:
#> ff
#> DMTA_M23 0.10705
#> DMTA_M27 0.09417
#> DMTA_M31 0.08919
#> DMTA_sink 0.70959
#>
#> Estimated disappearance times:
#> DT50 DT90 DT50back DT50_k1 DT50_k2
#> DMTA 13.81 49.3 14.84 12.91 49.85
#> M23 55.85 185.5 NA NA NA
#> M27 41.36 137.4 NA NA NA
#> M31 42.87 142.4 NA NA NA
plot(f_dmta_nlmixr_saem)
# Using saemix takes about 18 minutes
system.time(
f_dmta_saemix <- saem(f_dmta_mkin_tc, test_log_parms = TRUE)
)
#> DINTDY- T (=R1) illegal
#> In above message, R1 = 115.507
#>
#> T not in interval TCUR - HU (= R1) to TCUR (=R2)
#> In above message, R1 = 112.133, R2 = 113.577
#>
#> DLSODA- At T (=R1), too much accuracy requested
#> for precision of machine.. See TOLSF (=R2)
#> In above message, R1 = 55.3899, R2 = nan
#>
#> Error in out[available, var]: (subscript) logical subscript too long
#> Timing stopped at: 12.58 0 12.58
#> Timing stopped at: 12.99 0.008 13
# nlmixr with est = "saem" is pretty fast with default iteration numbers, most
# of the time (about 2.5 minutes) is spent for calculating the log likelihood at the end
# The likelihood calculated for the nlmixr fit is much lower than that found by saemix
# Also, the trace plot and the plot of the individual predictions is not
# convincing for the parent. It seems we are fitting an overparameterised
# model, so the result we get strongly depends on starting parameters and control settings.
system.time(
f_dmta_nlmixr_saem <- nlmixr(f_dmta_mkin_tc, est = "saem",
control = nlmixr::saemControl(print = 500, logLik = TRUE, nmc = 9))
)
#> With est = 'saem', a different error model is required for each observed variableChanging the error model to 'obs_tc' (Two-component error for each observed variable)
#> ℹ parameter labels from comments are typically ignored in non-interactive mode
#> ℹ Need to run with the source intact to parse comments
#>
#> → generate SAEM model
#> ✔ done
#> 1: 98.7179 -3.4492 -3.2592 -3.6952 -2.1629 -2.7824 0.8990 -2.8080 -2.7380 -2.8041 2.7789 0.6848 0.8170 0.7125 0.8550 1.5200 2.9882 0.3073 0.2850 0.2877 4.0480 0.4153 4.5214 0.3775 4.4419 0.4181 3.7069 0.5935
#> 500: 97.8519 -4.3891 -4.0888 -4.1247 -2.9246 -4.2755 2.6294 -2.1212 -2.1380 -2.0739 3.1293 1.2665 0.2763 0.3429 0.5743 1.5561 4.4991 0.1499 0.1551 0.3103 0.9514 0.0341 0.4846 0.1068 0.6597 0.0767 0.7836 0.0360
#> Calculating covariance matrix
#>
#> Calculating -2LL by Gaussian quadrature (nnodes=3,nsd=1.6)
#>
#> → creating full model...
#> → pruning branches (`if`/`else`)...
#> ✔ done
#> → loading into symengine environment...
#> ✔ done
#> → compiling EBE model...
#>
#> ✔ done
#> Needed Covariates:
#> [1] "CMT"
#> Calculating residuals/tables
#> done
#> user system elapsed
#> 785.825 3.841 153.598
traceplot(f_dmta_nlmixr_saem$nm)
#> Error in traceplot(f_dmta_nlmixr_saem$nm): could not find function "traceplot"
summary(f_dmta_nlmixr_saem)
#> nlmixr version used for fitting: 2.0.6
#> mkin version used for pre-fitting: 1.1.0
#> R version used for fitting: 4.1.2
#> Date of fit: Wed Mar 2 13:30:09 2022
#> Date of summary: Wed Mar 2 13:30:09 2022
#>
#> Equations:
#> d_DMTA/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 *
#> time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time)))
#> * DMTA
#> d_M23/dt = + f_DMTA_to_M23 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g)
#> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
#> exp(-k2 * time))) * DMTA - k_M23 * M23
#> d_M27/dt = + f_DMTA_to_M27 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g)
#> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
#> exp(-k2 * time))) * DMTA - k_M27 * M27 + k_M31 * M31
#> d_M31/dt = + f_DMTA_to_M31 * ((k1 * g * exp(-k1 * time) + k2 * (1 - g)
#> * exp(-k2 * time)) / (g * exp(-k1 * time) + (1 - g) *
#> exp(-k2 * time))) * DMTA - k_M31 * M31
#>
#> Data:
#> 563 observations of 4 variable(s) grouped in 6 datasets
#>
#> Degradation model predictions using RxODE
#>
#> Fitted in 153.313 s
#>
#> Variance model: Two-component variance function
#>
#> Mean of starting values for individual parameters:
#> DMTA_0 log_k_M23 log_k_M27 log_k_M31 f_DMTA_ilr_1 f_DMTA_ilr_2
#> 98.7132 -3.9216 -4.3306 -4.2442 0.1376 0.1388
#> f_DMTA_ilr_3 log_k1 log_k2 g_qlogis
#> -1.7554 -2.2352 -3.7758 0.4363
#>
#> Mean of starting values for error model parameters:
#> sigma_low_DMTA rsd_high_DMTA sigma_low_M23 rsd_high_M23 sigma_low_M27
#> 0.70012 0.02577 0.70012 0.02577 0.70012
#> rsd_high_M27 sigma_low_M31 rsd_high_M31
#> 0.02577 0.70012 0.02577
#>
#> Fixed degradation parameter values:
#> None
#>
#> Results:
#>
#> Likelihood calculated by focei
#> AIC BIC logLik
#> 1966 2088 -955.2
#>
#> Optimised parameters:
#> est. lower upper
#> DMTA_0 97.852 95.86386 99.840
#> log_k_M23 -4.389 -5.35084 -3.427
#> log_k_M27 -4.089 -4.54432 -3.633
#> log_k_M31 -4.125 -4.63280 -3.617
#> log_k1 -2.925 -3.54158 -2.308
#> log_k2 -4.275 -5.81760 -2.733
#> g_qlogis 2.629 -0.01785 5.277
#> f_DMTA_tffm0_1_qlogis -2.121 -2.44462 -1.798
#> f_DMTA_tffm0_2_qlogis -2.138 -2.47804 -1.798
#> f_DMTA_tffm0_3_qlogis -2.074 -2.53581 -1.612
#>
#> Correlation:
#> DMTA_0 lg__M23 lg__M27 lg__M31 log_k1 log_k2 g_qlogs
#> log_k_M23 -0.0164
#> log_k_M27 -0.0267 0.0028
#> log_k_M31 -0.0179 0.0023 0.0755
#> log_k1 0.0385 -0.0034 -0.0054 -0.0029
#> log_k2 0.0381 0.0115 0.0087 0.0093 0.0786
#> g_qlogis -0.0656 0.0021 0.0051 0.0001 -0.1177 -0.4389
#> f_DMTA_tffm0_1_qlogis -0.0604 0.0554 0.0054 0.0039 -0.0082 -0.0022 0.0119
#> f_DMTA_tffm0_2_qlogis -0.0601 0.0091 0.0577 -0.0350 -0.0081 -0.0057 0.0137
#> f_DMTA_tffm0_3_qlogis -0.0515 0.0083 0.0569 0.0729 -0.0059 0.0005 0.0073
#> f_DMTA_0_1 f_DMTA_0_2
#> log_k_M23
#> log_k_M27
#> log_k_M31
#> log_k1
#> log_k2
#> g_qlogis
#> f_DMTA_tffm0_1_qlogis
#> f_DMTA_tffm0_2_qlogis 0.0167
#> f_DMTA_tffm0_3_qlogis 0.0145 -0.0060
#>
#> Random effects (omega):
#> eta.DMTA_0 eta.log_k_M23 eta.log_k_M27 eta.log_k_M31
#> eta.DMTA_0 3.129 0.000 0.0000 0.0000
#> eta.log_k_M23 0.000 1.266 0.0000 0.0000
#> eta.log_k_M27 0.000 0.000 0.2763 0.0000
#> eta.log_k_M31 0.000 0.000 0.0000 0.3429
#> eta.log_k1 0.000 0.000 0.0000 0.0000
#> eta.log_k2 0.000 0.000 0.0000 0.0000
#> eta.g_qlogis 0.000 0.000 0.0000 0.0000
#> eta.f_DMTA_tffm0_1_qlogis 0.000 0.000 0.0000 0.0000
#> eta.f_DMTA_tffm0_2_qlogis 0.000 0.000 0.0000 0.0000
#> eta.f_DMTA_tffm0_3_qlogis 0.000 0.000 0.0000 0.0000
#> eta.log_k1 eta.log_k2 eta.g_qlogis
#> eta.DMTA_0 0.0000 0.000 0.000
#> eta.log_k_M23 0.0000 0.000 0.000
#> eta.log_k_M27 0.0000 0.000 0.000
#> eta.log_k_M31 0.0000 0.000 0.000
#> eta.log_k1 0.5743 0.000 0.000
#> eta.log_k2 0.0000 1.556 0.000
#> eta.g_qlogis 0.0000 0.000 4.499
#> eta.f_DMTA_tffm0_1_qlogis 0.0000 0.000 0.000
#> eta.f_DMTA_tffm0_2_qlogis 0.0000 0.000 0.000
#> eta.f_DMTA_tffm0_3_qlogis 0.0000 0.000 0.000
#> eta.f_DMTA_tffm0_1_qlogis eta.f_DMTA_tffm0_2_qlogis
#> eta.DMTA_0 0.0000 0.0000
#> eta.log_k_M23 0.0000 0.0000
#> eta.log_k_M27 0.0000 0.0000
#> eta.log_k_M31 0.0000 0.0000
#> eta.log_k1 0.0000 0.0000
#> eta.log_k2 0.0000 0.0000
#> eta.g_qlogis 0.0000 0.0000
#> eta.f_DMTA_tffm0_1_qlogis 0.1499 0.0000
#> eta.f_DMTA_tffm0_2_qlogis 0.0000 0.1551
#> eta.f_DMTA_tffm0_3_qlogis 0.0000 0.0000
#> eta.f_DMTA_tffm0_3_qlogis
#> eta.DMTA_0 0.0000
#> eta.log_k_M23 0.0000
#> eta.log_k_M27 0.0000
#> eta.log_k_M31 0.0000
#> eta.log_k1 0.0000
#> eta.log_k2 0.0000
#> eta.g_qlogis 0.0000
#> eta.f_DMTA_tffm0_1_qlogis 0.0000
#> eta.f_DMTA_tffm0_2_qlogis 0.0000
#> eta.f_DMTA_tffm0_3_qlogis 0.3103
#>
#> Variance model:
#> sigma_low_DMTA rsd_high_DMTA sigma_low_M23 rsd_high_M23 sigma_low_M27
#> 0.95135 0.03412 0.48455 0.10682 0.65969
#> rsd_high_M27 sigma_low_M31 rsd_high_M31
#> 0.07670 0.78365 0.03598
#>
#> Backtransformed parameters:
#> est. lower upper
#> DMTA_0 97.85189 95.863863 99.83992
#> k_M23 0.01241 0.004744 0.03247
#> k_M27 0.01676 0.010627 0.02643
#> k_M31 0.01617 0.009727 0.02687
#> f_DMTA_to_M23 0.10705 NA NA
#> f_DMTA_to_M27 0.09417 NA NA
#> f_DMTA_to_M31 0.08919 NA NA
#> k1 0.05369 0.028968 0.09950
#> k2 0.01391 0.002975 0.06500
#> g 0.93273 0.495538 0.99492
#>
#> Resulting formation fractions:
#> ff
#> DMTA_M23 0.10705
#> DMTA_M27 0.09417
#> DMTA_M31 0.08919
#> DMTA_sink 0.70959
#>
#> Estimated disappearance times:
#> DT50 DT90 DT50back DT50_k1 DT50_k2
#> DMTA 13.81 49.3 14.84 12.91 49.85
#> M23 55.85 185.5 NA NA NA
#> M27 41.36 137.4 NA NA NA
#> M31 42.87 142.4 NA NA NA
plot(f_dmta_nlmixr_saem)
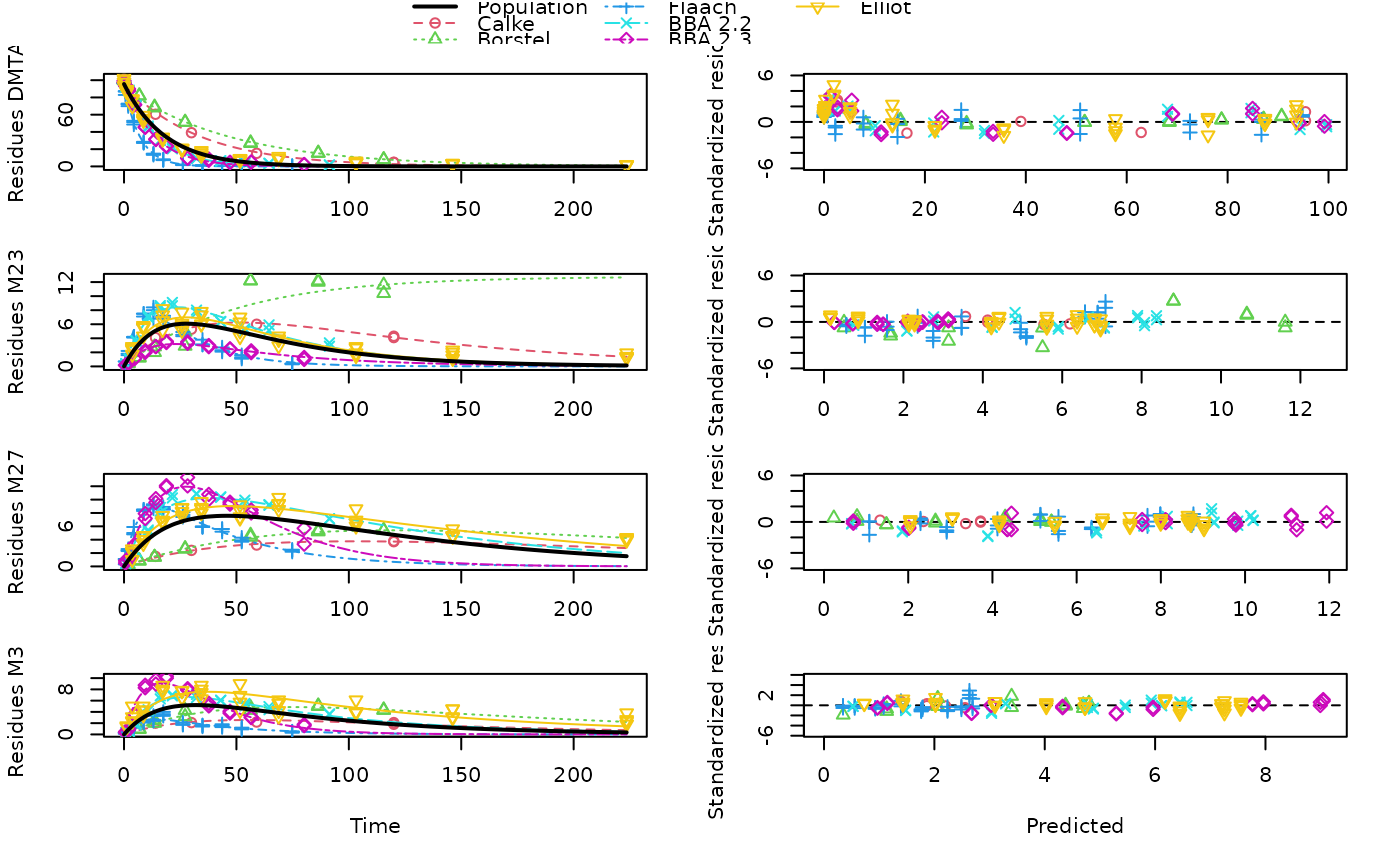 # }
# }