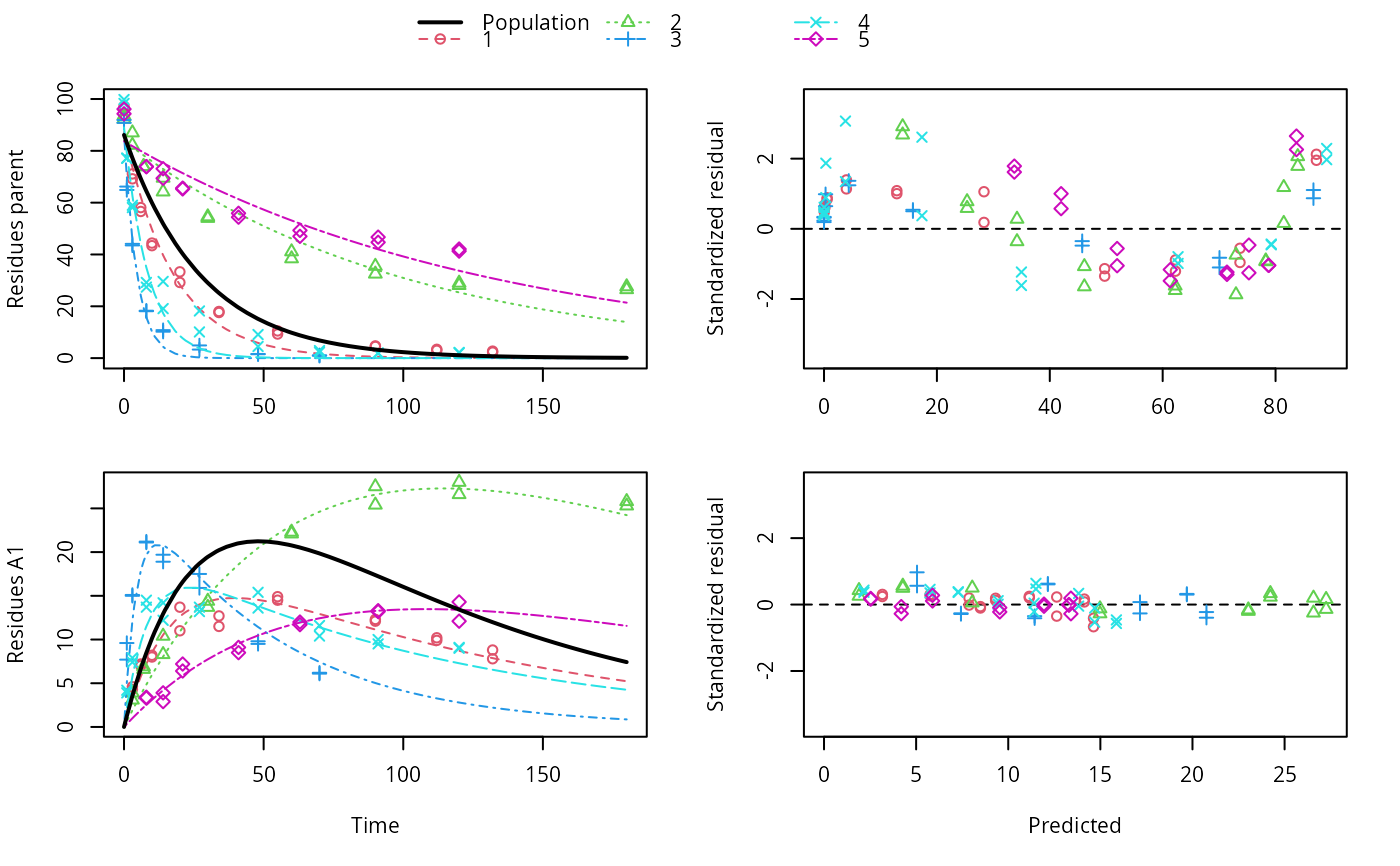This functions sets up a nonlinear mixed effects model for an mmkin row object. An mmkin row object is essentially a list of mkinfit objects that have been obtained by fitting the same model to a list of datasets.
# S3 method for mmkin nlme( model, data = "auto", fixed = lapply(as.list(names(mean_degparms(model))), function(el) eval(parse(text = paste(el, 1, sep = "~")))), random = pdDiag(fixed), groups, start = mean_degparms(model, random = TRUE), correlation = NULL, weights = NULL, subset, method = c("ML", "REML"), na.action = na.fail, naPattern, control = list(), verbose = FALSE ) # S3 method for nlme.mmkin print(x, digits = max(3, getOption("digits") - 3), ...) # S3 method for nlme.mmkin update(object, ...)
Arguments
| model | An mmkin row object. |
|---|---|
| data | Ignored, data are taken from the mmkin model |
| fixed | Ignored, all degradation parameters fitted in the mmkin model are used as fixed parameters |
| random | If not specified, correlated random effects are set up for all optimised degradation model parameters using the log-Cholesky parameterization nlme::pdLogChol that is also the default of the generic nlme method. |
| groups | See the documentation of nlme |
| start | If not specified, mean values of the fitted degradation parameters taken from the mmkin object are used |
| correlation | See the documentation of nlme |
| weights | passed to nlme |
| subset | passed to nlme |
| method | passed to nlme |
| na.action | passed to nlme |
| naPattern | passed to nlme |
| control | passed to nlme |
| verbose | passed to nlme |
| x | An nlme.mmkin object to print |
| digits | Number of digits to use for printing |
| ... | Update specifications passed to update.nlme |
| object | An nlme.mmkin object to update |
Value
Upon success, a fitted 'nlme.mmkin' object, which is an nlme object with additional elements. It also inherits from 'mixed.mmkin'.
Note
As the object inherits from nlme::nlme, there is a wealth of
methods that will automatically work on 'nlme.mmkin' objects, such as
nlme::intervals(), nlme::anova.lme() and nlme::coef.lme().
See also
Examples
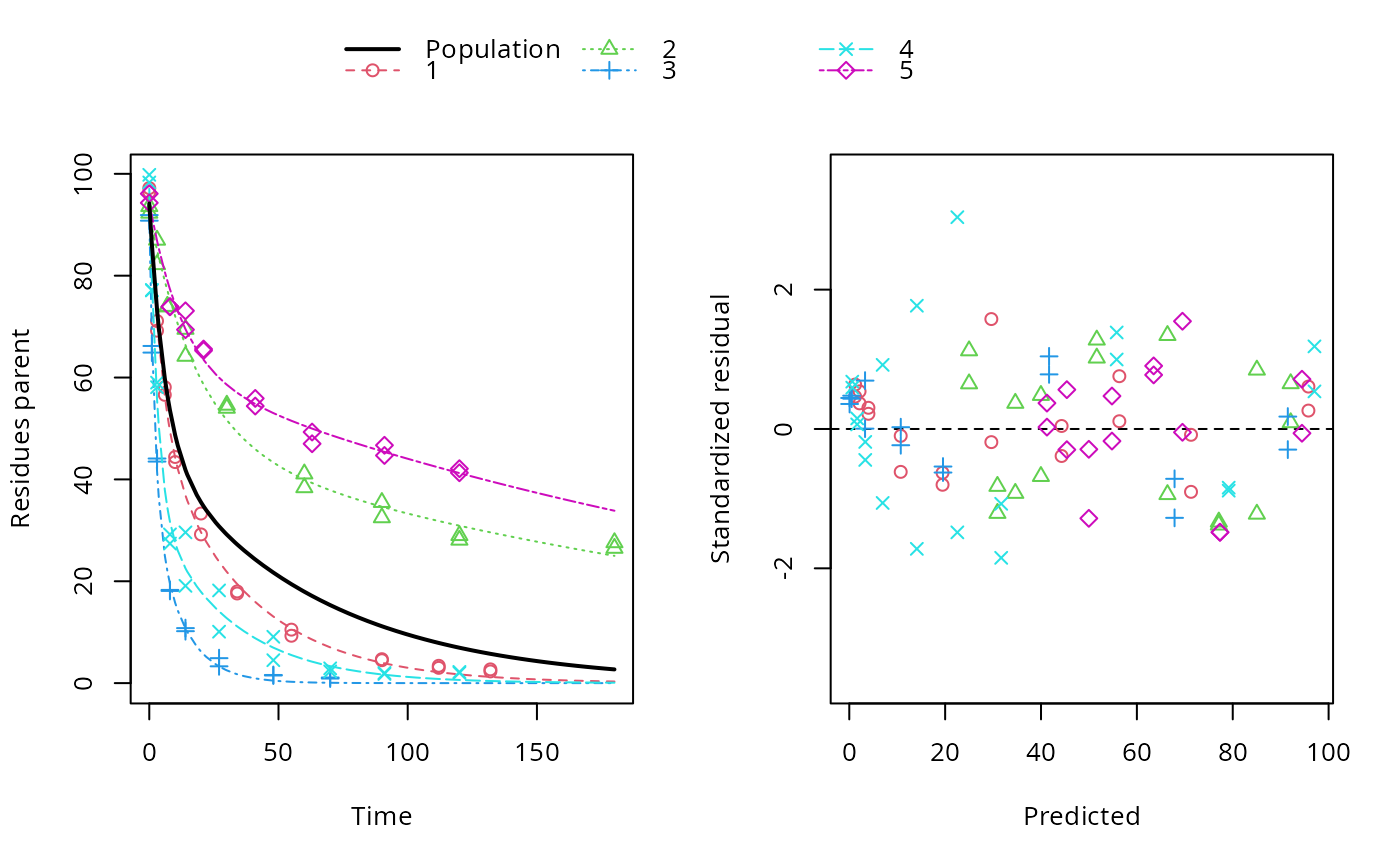
ds <- lapply(experimental_data_for_UBA_2019[6:10], function(x) subset(x$data[c("name", "time", "value")], name == "parent")) f <- mmkin(c("SFO", "DFOP"), ds, quiet = TRUE, cores = 1) library(nlme) f_nlme_sfo <- nlme(f["SFO", ]) # \dontrun{ f_nlme_dfop <- nlme(f["DFOP", ]) anova(f_nlme_sfo, f_nlme_dfop)#> Model df AIC BIC logLik Test L.Ratio p-value #> f_nlme_sfo 1 5 625.0539 637.5529 -307.5269 #> f_nlme_dfop 2 9 495.1270 517.6253 -238.5635 1 vs 2 137.9268 <.0001#> Kinetic nonlinear mixed-effects model fit by maximum likelihood #> #> Structural model: #> d_parent/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 * #> time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time))) #> * parent #> #> Data: #> 90 observations of 1 variable(s) grouped in 5 datasets #> #> Log-likelihood: -238.6 #> #> Fixed effects: #> list(parent_0 ~ 1, log_k1 ~ 1, log_k2 ~ 1, g_qlogis ~ 1) #> parent_0 log_k1 log_k2 g_qlogis #> 94.1702 -1.8002 -4.1474 0.0324 #> #> Random effects: #> Formula: list(parent_0 ~ 1, log_k1 ~ 1, log_k2 ~ 1, g_qlogis ~ 1) #> Level: ds #> Structure: Diagonal #> parent_0 log_k1 log_k2 g_qlogis Residual #> StdDev: 2.488 0.8447 1.33 0.4652 2.321 #>#> $distimes #> DT50 DT90 DT50back DT50_k1 DT50_k2 #> parent 10.79857 100.7937 30.34192 4.193937 43.85442 #>ds_2 <- lapply(experimental_data_for_UBA_2019[6:10], function(x) x$data[c("name", "time", "value")]) m_sfo_sfo <- mkinmod(parent = mkinsub("SFO", "A1"), A1 = mkinsub("SFO"), use_of_ff = "min", quiet = TRUE) m_sfo_sfo_ff <- mkinmod(parent = mkinsub("SFO", "A1"), A1 = mkinsub("SFO"), use_of_ff = "max", quiet = TRUE) m_dfop_sfo <- mkinmod(parent = mkinsub("DFOP", "A1"), A1 = mkinsub("SFO"), quiet = TRUE) f_2 <- mmkin(list("SFO-SFO" = m_sfo_sfo, "SFO-SFO-ff" = m_sfo_sfo_ff, "DFOP-SFO" = m_dfop_sfo), ds_2, quiet = TRUE) f_nlme_sfo_sfo <- nlme(f_2["SFO-SFO", ]) plot(f_nlme_sfo_sfo)# With formation fractions this does not coverge with defaults # f_nlme_sfo_sfo_ff <- nlme(f_2["SFO-SFO-ff", ]) #plot(f_nlme_sfo_sfo_ff) # With the log-Cholesky parameterization, this converges in 11 # iterations and around 100 seconds, but without tweaking control # parameters (with pdDiag, increasing the tolerance and pnlsMaxIter was # necessary) f_nlme_dfop_sfo <- nlme(f_2["DFOP-SFO", ])#> Error in nlme.formula(model = value ~ (mkin::get_deg_func())(name, time, parent_0, log_k_A1, f_parent_qlogis, log_k1, log_k2, g_qlogis), data = structure(list(ds = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L ), .Label = c("1", "2", "3", "4", "5"), class = c("ordered", "factor")), name = c("parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1"), time = c(0, 0, 3, 3, 6, 6, 10, 10, 20, 20, 34, 34, 55, 55, 90, 90, 112, 112, 132, 132, 3, 3, 6, 6, 10, 10, 20, 20, 34, 34, 55, 55, 90, 90, 112, 112, 132, 132, 0, 0, 3, 3, 7, 7, 14, 14, 30, 30, 60, 60, 90, 90, 120, 120, 180, 180, 3, 3, 7, 7, 14, 14, 30, 30, 60, 60, 90, 90, 120, 120, 180, 180, 0, 0, 1, 1, 3, 3, 8, 8, 14, 14, 27, 27, 48, 48, 70, 70, 1, 1, 3, 3, 8, 8, 14, 14, 27, 27, 48, 48, 70, 70, 0, 0, 1, 1, 3, 3, 8, 8, 14, 14, 27, 27, 48, 48, 70, 70, 91, 91, 120, 120, 1, 1, 3, 3, 8, 8, 14, 14, 27, 27, 48, 48, 70, 70, 91, 91, 120, 120, 0, 0, 8, 8, 14, 14, 21, 21, 41, 41, 63, 63, 91, 91, 120, 120, 8, 8, 14, 14, 21, 21, 41, 41, 63, 63, 91, 91, 120, 120), value = c(97.2, 96.4, 71.1, 69.2, 58.1, 56.6, 44.4, 43.4, 33.3, 29.2, 17.6, 18, 10.5, 9.3, 4.5, 4.7, 3, 3.4, 2.3, 2.7, 4.3, 4.6, 7, 7.2, 8.2, 8, 11, 13.7, 11.5, 12.7, 14.9, 14.5, 12.1, 12.3, 9.9, 10.2, 8.8, 7.8, 93.6, 92.3, 87, 82.2, 74, 73.9, 64.2, 69.5, 54, 54.6, 41.1, 38.4, 32.5, 35.5, 28.1, 29, 26.5, 27.6, 3.9, 3.1, 6.9, 6.6, 10.4, 8.3, 14.4, 13.7, 22.1, 22.3, 27.5, 25.4, 28, 26.6, 25.8, 25.3, 91.9, 90.8, 64.9, 66.2, 43.5, 44.1, 18.3, 18.1, 10.2, 10.8, 4.9, 3.3, 1.6, 1.5, 1.1, 0.9, 9.6, 7.7, 15, 15.1, 21.2, 21.1, 19.7, 18.9, 17.5, 15.9, 9.5, 9.8, 6.2, 6.1, 99.8, 98.3, 77.1, 77.2, 59, 58.1, 27.4, 29.2, 19.1, 29.6, 10.1, 18.2, 4.5, 9.1, 2.3, 2.9, 2, 1.8, 2, 2.2, 4.2, 3.9, 7.4, 7.9, 14.5, 13.7, 14.2, 12.2, 13.7, 13.2, 13.6, 15.4, 10.4, 11.6, 10, 9.5, 9.1, 9, 96.1, 94.3, 73.9, 73.9, 69.4, 73.1, 65.6, 65.3, 55.9, 54.4, 47, 49.3, 44.7, 46.7, 42.1, 41.3, 3.3, 3.4, 3.9, 2.9, 6.4, 7.2, 9.1, 8.5, 11.7, 12, 13.3, 13.2, 14.3, 12.1)), row.names = c(NA, -170L), class = c("nfnGroupedData", "nfGroupedData", "groupedData", "data.frame"), formula = value ~ time | ds, FUN = function (x) max(x, na.rm = TRUE), order.groups = FALSE), start = list( fixed = c(parent_0 = 93.8101519326534, log_k_A1 = -9.76474551635931, f_parent_qlogis = -0.971114801595408, log_k1 = -1.87993711571859, log_k2 = -4.27081421366622, g_qlogis = 0.135644115277507 ), random = list(ds = structure(c(2.56569977430371, -3.49441920289139, -3.32614443321494, 4.35347873814922, -0.0986148763466161, 4.65850590018027, 1.8618544764481, 6.12693257601545, 4.91792724701579, -17.5652201996596, -0.466203822618637, 0.746660653597927, 0.282193987271096, -0.42053488943072, -0.142115928819667, 0.369240076779088, -1.38985563501659, 1.02592753494098, 0.73090914081534, -0.736221117518819, 0.768170629350299, -1.89347658079869, 1.72168783460352, 0.844607177798114, -1.44098906095325, -0.377731855445672, 0.168180098477565, 0.469683412912104, 0.500717664434525, -0.760849320378522), .Dim = 5:6, .Dimnames = list(c("1", "2", "3", "4", "5"), c("parent_0", "log_k_A1", "f_parent_qlogis", "log_k1", "log_k2", "g_qlogis"))))), fixed = list(parent_0 ~ 1, log_k_A1 ~ 1, f_parent_qlogis ~ 1, log_k1 ~ 1, log_k2 ~ 1, g_qlogis ~ 1), random = structure(numeric(0), class = c("pdDiag", "pdMat"), formula = structure(list(parent_0 ~ 1, log_k_A1 ~ 1, f_parent_qlogis ~ 1, log_k1 ~ 1, log_k2 ~ 1, g_qlogis ~ 1), class = "listForm"), Dimnames = list(NULL, NULL))): maximum number of iterations (maxIter = 50) reached without convergence#>#> Error in plot(f_nlme_dfop_sfo): object 'f_nlme_dfop_sfo' not found#> Error in anova(f_nlme_dfop_sfo, f_nlme_sfo_sfo): object 'f_nlme_dfop_sfo' not found#> $ff #> parent_sink parent_A1 A1_sink #> 0.5912432 0.4087568 1.0000000 #> #> $distimes #> DT50 DT90 #> parent 19.13518 63.5657 #> A1 66.02155 219.3189 #>#> Error in endpoints(f_nlme_dfop_sfo): object 'f_nlme_dfop_sfo' not foundif (length(findFunction("varConstProp")) > 0) { # tc error model for nlme available # Attempts to fit metabolite kinetics with the tc error model are possible, # but need tweeking of control values and sometimes do not converge f_tc <- mmkin(c("SFO", "DFOP"), ds, quiet = TRUE, error_model = "tc") f_nlme_sfo_tc <- nlme(f_tc["SFO", ]) f_nlme_dfop_tc <- nlme(f_tc["DFOP", ]) AIC(f_nlme_sfo, f_nlme_sfo_tc, f_nlme_dfop, f_nlme_dfop_tc) print(f_nlme_dfop_tc) }#> Kinetic nonlinear mixed-effects model fit by maximum likelihood #> #> Structural model: #> d_parent/dt = - ((k1 * g * exp(-k1 * time) + k2 * (1 - g) * exp(-k2 * #> time)) / (g * exp(-k1 * time) + (1 - g) * exp(-k2 * time))) #> * parent #> #> Data: #> 90 observations of 1 variable(s) grouped in 5 datasets #> #> Log-likelihood: -238.4 #> #> Fixed effects: #> list(parent_0 ~ 1, log_k1 ~ 1, log_k2 ~ 1, g_qlogis ~ 1) #> parent_0 log_k1 log_k2 g_qlogis #> 94.04775 -1.82340 -4.16715 0.05685 #> #> Random effects: #> Formula: list(parent_0 ~ 1, log_k1 ~ 1, log_k2 ~ 1, g_qlogis ~ 1) #> Level: ds #> Structure: Diagonal #> parent_0 log_k1 log_k2 g_qlogis Residual #> StdDev: 2.474 0.85 1.337 0.4659 1 #> #> Variance function: #> Structure: Constant plus proportion of variance covariate #> Formula: ~fitted(.) #> Parameter estimates: #> const prop #> 2.23224114 0.01262341f_2_obs <- mmkin(list("SFO-SFO" = m_sfo_sfo, "DFOP-SFO" = m_dfop_sfo), ds_2, quiet = TRUE, error_model = "obs") f_nlme_sfo_sfo_obs <- nlme(f_2_obs["SFO-SFO", ]) print(f_nlme_sfo_sfo_obs)#> Kinetic nonlinear mixed-effects model fit by maximum likelihood #> #> Structural model: #> d_parent/dt = - k_parent_sink * parent - k_parent_A1 * parent #> d_A1/dt = + k_parent_A1 * parent - k_A1_sink * A1 #> #> Data: #> 170 observations of 2 variable(s) grouped in 5 datasets #> #> Log-likelihood: -473 #> #> Fixed effects: #> list(parent_0 ~ 1, log_k_parent_sink ~ 1, log_k_parent_A1 ~ 1, log_k_A1_sink ~ 1) #> parent_0 log_k_parent_sink log_k_parent_A1 log_k_A1_sink #> 87.976 -3.670 -4.164 -4.645 #> #> Random effects: #> Formula: list(parent_0 ~ 1, log_k_parent_sink ~ 1, log_k_parent_A1 ~ 1, log_k_A1_sink ~ 1) #> Level: ds #> Structure: Diagonal #> parent_0 log_k_parent_sink log_k_parent_A1 log_k_A1_sink Residual #> StdDev: 3.992 1.777 1.055 0.4821 6.483 #> #> Variance function: #> Structure: Different standard deviations per stratum #> Formula: ~1 | name #> Parameter estimates: #> parent A1 #> 1.0000000 0.2050003#> Error in nlme.formula(model = value ~ (mkin::get_deg_func())(name, time, parent_0, log_k_A1, f_parent_qlogis, log_k1, log_k2, g_qlogis), data = structure(list(ds = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L ), .Label = c("1", "2", "3", "4", "5"), class = c("ordered", "factor")), name = c("parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "parent", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1"), time = c(0, 0, 3, 3, 6, 6, 10, 10, 20, 20, 34, 34, 55, 55, 90, 90, 112, 112, 132, 132, 3, 3, 6, 6, 10, 10, 20, 20, 34, 34, 55, 55, 90, 90, 112, 112, 132, 132, 0, 0, 3, 3, 7, 7, 14, 14, 30, 30, 60, 60, 90, 90, 120, 120, 180, 180, 3, 3, 7, 7, 14, 14, 30, 30, 60, 60, 90, 90, 120, 120, 180, 180, 0, 0, 1, 1, 3, 3, 8, 8, 14, 14, 27, 27, 48, 48, 70, 70, 1, 1, 3, 3, 8, 8, 14, 14, 27, 27, 48, 48, 70, 70, 0, 0, 1, 1, 3, 3, 8, 8, 14, 14, 27, 27, 48, 48, 70, 70, 91, 91, 120, 120, 1, 1, 3, 3, 8, 8, 14, 14, 27, 27, 48, 48, 70, 70, 91, 91, 120, 120, 0, 0, 8, 8, 14, 14, 21, 21, 41, 41, 63, 63, 91, 91, 120, 120, 8, 8, 14, 14, 21, 21, 41, 41, 63, 63, 91, 91, 120, 120), value = c(97.2, 96.4, 71.1, 69.2, 58.1, 56.6, 44.4, 43.4, 33.3, 29.2, 17.6, 18, 10.5, 9.3, 4.5, 4.7, 3, 3.4, 2.3, 2.7, 4.3, 4.6, 7, 7.2, 8.2, 8, 11, 13.7, 11.5, 12.7, 14.9, 14.5, 12.1, 12.3, 9.9, 10.2, 8.8, 7.8, 93.6, 92.3, 87, 82.2, 74, 73.9, 64.2, 69.5, 54, 54.6, 41.1, 38.4, 32.5, 35.5, 28.1, 29, 26.5, 27.6, 3.9, 3.1, 6.9, 6.6, 10.4, 8.3, 14.4, 13.7, 22.1, 22.3, 27.5, 25.4, 28, 26.6, 25.8, 25.3, 91.9, 90.8, 64.9, 66.2, 43.5, 44.1, 18.3, 18.1, 10.2, 10.8, 4.9, 3.3, 1.6, 1.5, 1.1, 0.9, 9.6, 7.7, 15, 15.1, 21.2, 21.1, 19.7, 18.9, 17.5, 15.9, 9.5, 9.8, 6.2, 6.1, 99.8, 98.3, 77.1, 77.2, 59, 58.1, 27.4, 29.2, 19.1, 29.6, 10.1, 18.2, 4.5, 9.1, 2.3, 2.9, 2, 1.8, 2, 2.2, 4.2, 3.9, 7.4, 7.9, 14.5, 13.7, 14.2, 12.2, 13.7, 13.2, 13.6, 15.4, 10.4, 11.6, 10, 9.5, 9.1, 9, 96.1, 94.3, 73.9, 73.9, 69.4, 73.1, 65.6, 65.3, 55.9, 54.4, 47, 49.3, 44.7, 46.7, 42.1, 41.3, 3.3, 3.4, 3.9, 2.9, 6.4, 7.2, 9.1, 8.5, 11.7, 12, 13.3, 13.2, 14.3, 12.1)), row.names = c(NA, -170L), class = c("nfnGroupedData", "nfGroupedData", "groupedData", "data.frame"), formula = value ~ time | ds, FUN = function (x) max(x, na.rm = TRUE), order.groups = FALSE), start = list( fixed = c(parent_0 = 93.4272167134207, log_k_A1 = -9.71590717106959, f_parent_qlogis = -0.953712099744438, log_k1 = -1.95256957646888, log_k2 = -4.42919226610318, g_qlogis = 0.193023137298073 ), random = list(ds = structure(c(2.85557330683041, -3.87630303729395, -2.78062140212751, 4.82042042600536, -1.01906929341432, 4.613992019697, 2.05871276943309, 6.0766404049189, 4.86471337131288, -17.6140585653619, -0.480721175257541, 0.773079218835614, 0.260464433006093, -0.440615012802434, -0.112207463781733, 0.445812953745225, -1.49588630006094, 1.13602040717272, 0.801850880762046, -0.887797941619048, 0.936480292463262, -2.43093808171905, 1.91256225793793, 0.984827519864443, -1.40293198854659, -0.455176326336681, 0.376355651864385, 0.343919720700401, 0.46329187713133, -0.728390923359434 ), .Dim = 5:6, .Dimnames = list(c("1", "2", "3", "4", "5"), c("parent_0", "log_k_A1", "f_parent_qlogis", "log_k1", "log_k2", "g_qlogis"))))), fixed = list(parent_0 ~ 1, log_k_A1 ~ 1, f_parent_qlogis ~ 1, log_k1 ~ 1, log_k2 ~ 1, g_qlogis ~ 1), random = structure(numeric(0), class = c("pdDiag", "pdMat"), formula = structure(list(parent_0 ~ 1, log_k_A1 ~ 1, f_parent_qlogis ~ 1, log_k1 ~ 1, log_k2 ~ 1, g_qlogis ~ 1), class = "listForm"), Dimnames = list(NULL, NULL)), weights = structure(numeric(0), formula = ~1 | name, class = c("varIdent", "varFunc"))): maximum number of iterations (maxIter = 50) reached without convergence#>f_2_tc <- mmkin(list("SFO-SFO" = m_sfo_sfo, "DFOP-SFO" = m_dfop_sfo), ds_2, quiet = TRUE, error_model = "tc") # f_nlme_sfo_sfo_tc <- nlme(f_2_tc["SFO-SFO", ]) # stops with error message f_nlme_dfop_sfo_tc <- nlme(f_2_tc["DFOP-SFO", ])#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Warning: longer object length is not a multiple of shorter object length#> Error in X[, fmap[[nm]]] <- gradnm: number of items to replace is not a multiple of replacement length#># We get warnings about false convergence in the LME step in several iterations # but as the last such warning occurs in iteration 25 and we have 28 iterations # we can ignore these anova(f_nlme_dfop_sfo, f_nlme_dfop_sfo_obs, f_nlme_dfop_sfo_tc)#> Error in anova(f_nlme_dfop_sfo, f_nlme_dfop_sfo_obs, f_nlme_dfop_sfo_tc): object 'f_nlme_dfop_sfo' not found# }