Function describing the standard deviation of the measurement error in dependence of the measured value \(y\):
sigma_twocomp(y, sigma_low, rsd_high)
Arguments
| y | The magnitude of the observed value |
|---|---|
| sigma_low | The asymptotic minimum of the standard deviation for low observed values |
| rsd_high | The coefficient describing the increase of the standard deviation with the magnitude of the observed value |
Value
The standard deviation of the response variable.
Details
$$\sigma = \sqrt{ \sigma_{low}^2 + y^2 * {rsd}_{high}^2}$$ sigma = sqrt(sigma_low^2 + y^2 * rsd_high^2)
This is the error model used for example by Werner et al. (1978). The model proposed by Rocke and Lorenzato (1995) can be written in this form as well, but assumes approximate lognormal distribution of errors for high values of y.
References
Werner, Mario, Brooks, Samuel H., and Knott, Lancaster B. (1978) Additive, Multiplicative, and Mixed Analytical Errors. Clinical Chemistry 24(11), 1895-1898.
Rocke, David M. and Lorenzato, Stefan (1995) A two-component model for measurement error in analytical chemistry. Technometrics 37(2), 176-184.
Examples
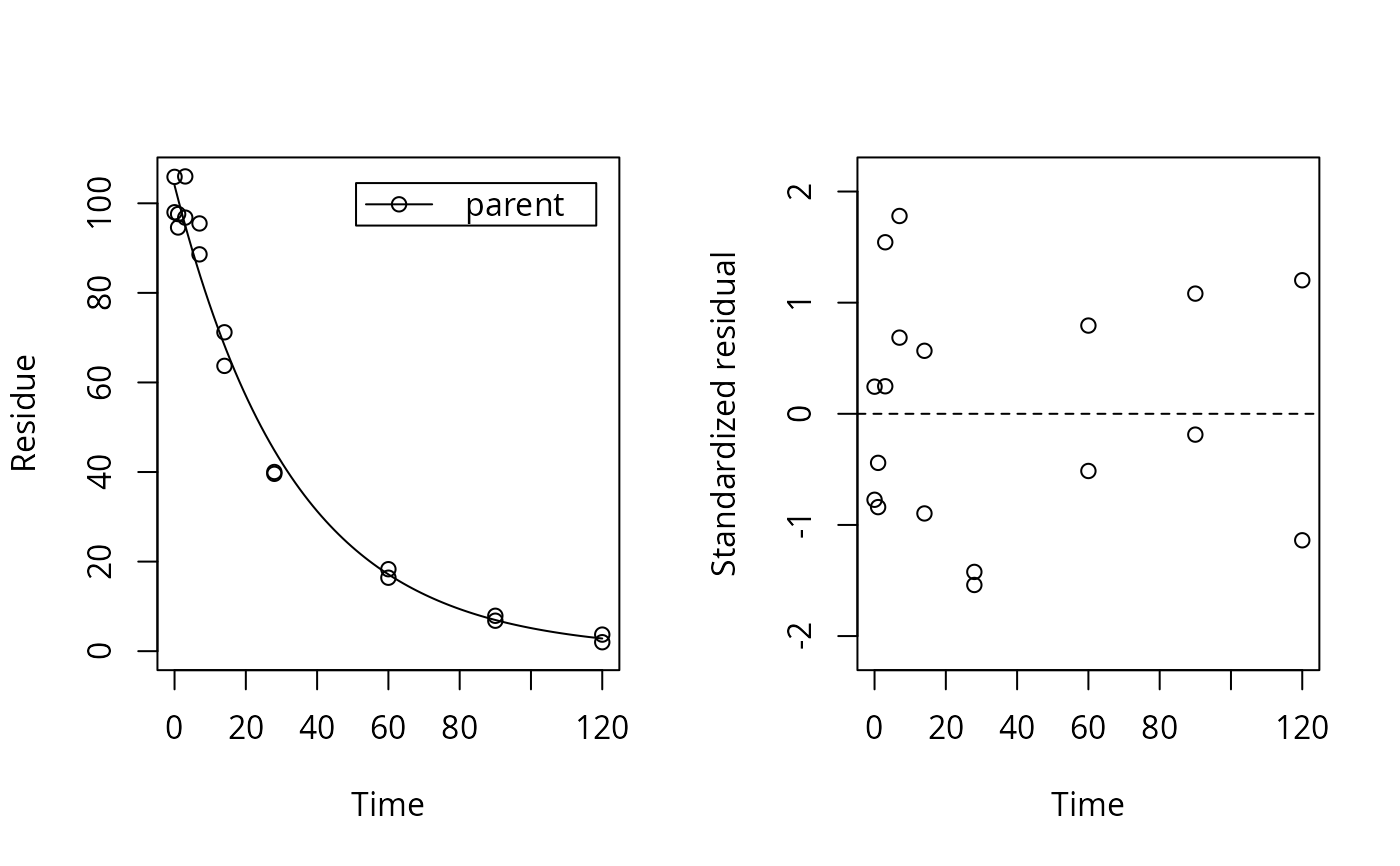
times <- c(0, 1, 3, 7, 14, 28, 60, 90, 120) d_pred <- data.frame(time = times, parent = 100 * exp(- 0.03 * times)) set.seed(123456) d_syn <- add_err(d_pred, function(y) sigma_twocomp(y, 1, 0.07), reps = 2, n = 1)[[1]] f_nls <- nls(value ~ SSasymp(time, 0, parent_0, lrc), data = d_syn, start = list(parent_0 = 100, lrc = -3)) library(nlme) f_gnls <- gnls(value ~ SSasymp(time, 0, parent_0, lrc), data = d_syn, na.action = na.omit, start = list(parent_0 = 100, lrc = -3)) if (length(findFunction("varConstProp")) > 0) { f_gnls_tc <- update(f_gnls, weights = varConstProp()) f_gnls_tc_sf <- update(f_gnls_tc, control = list(sigma = 1)) } f_mkin <- mkinfit("SFO", d_syn, error_model = "const", quiet = TRUE) f_mkin_tc <- mkinfit("SFO", d_syn, error_model = "tc", quiet = TRUE) plot_res(f_mkin_tc, standardized = TRUE)#> df AIC #> f_nls 3 114.4817 #> f_gnls 3 114.4817 #> f_gnls_tc 5 103.6447 #> f_gnls_tc_sf 4 101.6447 #> f_mkin 3 114.4817 #> f_mkin_tc 4 101.6446