Testing covariate modelling in hierarchical parent degradation kinetics with residue data on mesotrione
Johannes Ranke
Last change on 4 August 2023, last compiled on 16 Februar 2025
Source:vignettes/prebuilt/2023_mesotrione_parent.rmd
2023_mesotrione_parent.rmdIntroduction
The purpose of this document is to test demonstrate how nonlinear hierarchical models (NLHM) based on the parent degradation models SFO, FOMC, DFOP and HS can be fitted with the mkin package, also considering the influence of covariates like soil pH on different degradation parameters. Because in some other case studies, the SFORB parameterisation of biexponential decline has shown some advantages over the DFOP parameterisation, SFORB was included in the list of tested models as well.
The mkin package is used in version 1.2.10, which is contains the
functions that were used for the evaluations. The saemix
package is used as a backend for fitting the NLHM, but is also loaded to
make the convergence plot function available.
This document is processed with the knitr package, which
also provides the kable function that is used to improve
the display of tabular data in R markdown documents. For parallel
processing, the parallel package is used.
library(mkin)
library(knitr)
library(saemix)
library(parallel)
n_cores <- detectCores()
if (Sys.info()["sysname"] == "Windows") {
cl <- makePSOCKcluster(n_cores)
} else {
cl <- makeForkCluster(n_cores)
}Test data
data_file <- system.file(
"testdata", "mesotrione_soil_efsa_2016.xlsx", package = "mkin")
meso_ds <- read_spreadsheet(data_file, parent_only = TRUE)The following tables show the covariate data and the 18 datasets that were read in from the spreadsheet file.
| pH | |
|---|---|
| Richmond | 6.2 |
| Richmond 2 | 6.2 |
| ERTC | 6.4 |
| Toulouse | 7.7 |
| Picket Piece | 7.1 |
| 721 | 5.6 |
| 722 | 5.7 |
| 723 | 5.4 |
| 724 | 4.8 |
| 725 | 5.8 |
| 727 | 5.1 |
| 728 | 5.9 |
| 729 | 5.6 |
| 730 | 5.3 |
| 731 | 6.1 |
| 732 | 5.0 |
| 741 | 5.7 |
| 742 | 7.2 |
for (ds_name in names(meso_ds)) {
print(
kable(mkin_long_to_wide(meso_ds[[ds_name]]),
caption = paste("Dataset", ds_name),
booktabs = TRUE, row.names = FALSE))
}| time | meso |
|---|---|
| 0.000000 | 91.00 |
| 1.179050 | 86.70 |
| 3.537149 | 73.60 |
| 7.074299 | 61.50 |
| 10.611448 | 55.70 |
| 15.327647 | 47.70 |
| 17.685747 | 39.50 |
| 24.760046 | 29.80 |
| 35.371494 | 19.60 |
| 68.384889 | 5.67 |
| 0.000000 | 97.90 |
| 1.179050 | 96.40 |
| 3.537149 | 89.10 |
| 7.074299 | 74.40 |
| 10.611448 | 57.40 |
| 15.327647 | 46.30 |
| 18.864797 | 35.50 |
| 27.118146 | 27.20 |
| 35.371494 | 19.10 |
| 74.280138 | 6.50 |
| 108.472582 | 3.40 |
| 142.665027 | 2.20 |
| time | meso |
|---|---|
| 0.000000 | 96.0 |
| 2.422004 | 82.4 |
| 5.651343 | 71.2 |
| 8.073348 | 53.1 |
| 11.302687 | 48.5 |
| 16.954030 | 33.4 |
| 22.605373 | 24.2 |
| 45.210746 | 11.9 |
| time | meso |
|---|---|
| 0.000000 | 99.9 |
| 2.755193 | 80.0 |
| 6.428782 | 42.1 |
| 9.183975 | 50.1 |
| 12.857565 | 28.4 |
| 19.286347 | 39.8 |
| 25.715130 | 29.9 |
| 51.430259 | 2.5 |
| time | meso |
|---|---|
| 0.000000 | 96.8 |
| 2.897983 | 63.3 |
| 6.761960 | 22.3 |
| 9.659942 | 16.6 |
| 13.523919 | 16.1 |
| 20.285879 | 17.2 |
| 27.047838 | 1.8 |
| time | meso |
|---|---|
| 0.000000 | 102.0 |
| 2.841195 | 73.7 |
| 6.629454 | 35.5 |
| 9.470649 | 31.8 |
| 13.258909 | 18.0 |
| 19.888364 | 3.7 |
| time | meso |
|---|---|
| 0.00000 | 86.4 |
| 11.24366 | 61.4 |
| 22.48733 | 49.8 |
| 33.73099 | 41.0 |
| 44.97466 | 35.1 |
| time | meso |
|---|---|
| 0.00000 | 90.3 |
| 11.24366 | 52.1 |
| 22.48733 | 37.4 |
| 33.73099 | 21.2 |
| 44.97466 | 14.3 |
| time | meso |
|---|---|
| 0.00000 | 89.3 |
| 11.24366 | 70.8 |
| 22.48733 | 51.1 |
| 33.73099 | 42.7 |
| 44.97466 | 26.7 |
| time | meso |
|---|---|
| 0.000000 | 89.4 |
| 9.008208 | 65.2 |
| 18.016415 | 55.8 |
| 27.024623 | 46.0 |
| 36.032831 | 41.7 |
| time | meso |
|---|---|
| 0.00000 | 89.0 |
| 10.99058 | 35.4 |
| 21.98116 | 18.6 |
| 32.97174 | 11.6 |
| 43.96232 | 7.6 |
| time | meso |
|---|---|
| 0.00000 | 91.3 |
| 10.96104 | 63.2 |
| 21.92209 | 51.1 |
| 32.88313 | 42.0 |
| 43.84417 | 40.8 |
| time | meso |
|---|---|
| 0.00000 | 91.8 |
| 11.24366 | 43.6 |
| 22.48733 | 22.0 |
| 33.73099 | 15.9 |
| 44.97466 | 8.8 |
| time | meso |
|---|---|
| 0.00000 | 91.6 |
| 11.24366 | 60.5 |
| 22.48733 | 43.5 |
| 33.73099 | 28.4 |
| 44.97466 | 20.5 |
| time | meso |
|---|---|
| 0.00000 | 92.7 |
| 11.07446 | 58.9 |
| 22.14893 | 44.0 |
| 33.22339 | 46.0 |
| 44.29785 | 29.3 |
| time | meso |
|---|---|
| 0.00000 | 92.1 |
| 11.24366 | 64.4 |
| 22.48733 | 45.3 |
| 33.73099 | 33.6 |
| 44.97466 | 23.5 |
| time | meso |
|---|---|
| 0.00000 | 90.3 |
| 11.24366 | 58.2 |
| 22.48733 | 40.1 |
| 33.73099 | 33.1 |
| 44.97466 | 25.8 |
| time | meso |
|---|---|
| 0.00000 | 90.3 |
| 10.84712 | 68.7 |
| 21.69424 | 58.0 |
| 32.54136 | 52.2 |
| 43.38848 | 48.0 |
| time | meso |
|---|---|
| 0.00000 | 92.0 |
| 11.24366 | 60.9 |
| 22.48733 | 36.2 |
| 33.73099 | 18.3 |
| 44.97466 | 8.7 |
Separate evaluations
In order to obtain suitable starting parameters for the NLHM fits,
separate fits of the five models to the data for each soil are generated
using the mmkin function from the mkin package. In a first
step, constant variance is assumed. Convergence is checked with the
status function.
deg_mods <- c("SFO", "FOMC", "DFOP", "SFORB", "HS")
f_sep_const <- mmkin(
deg_mods,
meso_ds,
error_model = "const",
cluster = cl,
quiet = TRUE)| Richmond | Richmond 2 | ERTC | Toulouse | Picket Piece | |
|---|---|---|---|---|---|
| SFO | OK | OK | OK | OK | OK |
| FOMC | OK | OK | OK | OK | C |
| DFOP | OK | OK | OK | OK | OK |
| SFORB | OK | OK | OK | OK | OK |
| HS | OK | OK | C | OK | OK |
| 721 | 722 | 723 | 724 | 725 | 727 | 728 | 729 | 730 | 731 | 732 | 741 | 742 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SFO | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK |
| FOMC | OK | OK | C | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK |
| DFOP | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK |
| SFORB | OK | OK | OK | OK | OK | OK | OK | C | OK | OK | OK | OK | OK |
| HS | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK |
In the tables above, OK indicates convergence and C indicates failure to converge. Most separate fits with constant variance converged, with the exception of two FOMC fits, one SFORB fit and one HS fit.
f_sep_tc <- update(f_sep_const, error_model = "tc")| Richmond | Richmond 2 | ERTC | Toulouse | Picket Piece | |
|---|---|---|---|---|---|
| SFO | OK | OK | OK | OK | OK |
| FOMC | OK | OK | OK | OK | OK |
| DFOP | C | OK | OK | OK | OK |
| SFORB | OK | OK | OK | OK | OK |
| HS | OK | OK | C | OK | OK |
| 721 | 722 | 723 | 724 | 725 | 727 | 728 | 729 | 730 | 731 | 732 | 741 | 742 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SFO | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK | OK |
| FOMC | OK | OK | C | OK | C | C | OK | C | OK | C | OK | C | OK |
| DFOP | C | OK | OK | OK | C | OK | OK | OK | OK | C | OK | C | OK |
| SFORB | C | OK | OK | OK | C | OK | OK | C | OK | OK | OK | C | OK |
| HS | OK | OK | OK | OK | OK | OK | OK | OK | OK | C | OK | OK | OK |
With the two-component error model, the set of fits that did not converge is larger, with convergence problems appearing for a number of non-SFO fits.
Hierarchical model fits without covariate effect
The following code fits hierarchical kinetic models for the ten combinations of the five different degradation models with the two different error models in parallel.
| const | tc | |
|---|---|---|
| SFO | OK | OK |
| FOMC | OK | OK |
| DFOP | OK | OK |
| SFORB | OK | OK |
| HS | OK | OK |
All fits terminate without errors (status OK).
| npar | AIC | BIC | Lik | |
|---|---|---|---|---|
| SFO const | 5 | 800.0 | 804.5 | -395.0 |
| SFO tc | 6 | 801.9 | 807.2 | -394.9 |
| FOMC const | 7 | 787.4 | 793.6 | -386.7 |
| FOMC tc | 8 | 788.9 | 796.1 | -386.5 |
| DFOP const | 9 | 787.6 | 795.6 | -384.8 |
| SFORB const | 9 | 787.4 | 795.4 | -384.7 |
| HS const | 9 | 781.9 | 789.9 | -382.0 |
| DFOP tc | 10 | 787.4 | 796.3 | -383.7 |
| SFORB tc | 10 | 795.8 | 804.7 | -387.9 |
| HS tc | 10 | 783.7 | 792.7 | -381.9 |
The model comparisons show that the fits with constant variance are
consistently preferable to the corresponding fits with two-component
error for these data. This is confirmed by the fact that the parameter
b.1 (the relative standard deviation in the fits obtained
with the saemix package), is ill-defined in all fits.
| const | tc | |
|---|---|---|
| SFO | sd(meso_0) | sd(meso_0), b.1 |
| FOMC | sd(meso_0), sd(log_beta) | sd(meso_0), sd(log_beta), b.1 |
| DFOP | sd(meso_0), sd(log_k1) | sd(meso_0), sd(g_qlogis), b.1 |
| SFORB | sd(meso_free_0), sd(log_k_meso_free_bound) | sd(meso_free_0), sd(log_k_meso_free_bound), b.1 |
| HS | sd(meso_0) | sd(meso_0), b.1 |
For obtaining fits with only well-defined random effects, we update
the set of fits, excluding random effects that were ill-defined
according to the illparms function.
| const | tc | |
|---|---|---|
| SFO | OK | OK |
| FOMC | OK | OK |
| DFOP | OK | OK |
| SFORB | OK | OK |
| HS | OK | OK |
The updated fits terminate without errors.
| const | tc | |
|---|---|---|
| SFO | b.1 | |
| FOMC | b.1 | |
| DFOP | b.1 | |
| SFORB | b.1 | |
| HS | b.1 |
No ill-defined errors remain in the fits with constant variance.
Hierarchical model fits with covariate effect
In the following sections, hierarchical fits including a model for the influence of pH on selected degradation parameters are shown for all parent models. Constant variance is selected as the error model based on the fits without covariate effects. Random effects that were ill-defined in the fits without pH influence are excluded. A potential influence of the soil pH is only included for parameters with a well-defined random effect, because experience has shown that only for such parameters a significant pH effect could be found.
SFO
sfo_pH <- saem(f_sep_const["SFO", ], no_random_effect = "meso_0", covariates = pH,
covariate_models = list(log_k_meso ~ pH))| est. | lower | upper | |
|---|---|---|---|
| meso_0 | 91.35 | 89.27 | 93.43 |
| log_k_meso | -6.66 | -7.97 | -5.35 |
| beta_pH(log_k_meso) | 0.59 | 0.37 | 0.81 |
| a.1 | 5.48 | 4.71 | 6.24 |
| SD.log_k_meso | 0.35 | 0.23 | 0.47 |
The parameter showing the pH influence in the above table is
beta_pH(log_k_meso). Its confidence interval does not
include zero, indicating that the influence of soil pH on the log of the
degradation rate constant is significantly greater than zero.
anova(f_saem_2[["SFO", "const"]], sfo_pH, test = TRUE)Data: 116 observations of 1 variable(s) grouped in 18 datasets
npar AIC BIC Lik Chisq Df Pr(>Chisq)
f_saem_2[["SFO", "const"]] 4 797.56 801.12 -394.78
sfo_pH 5 783.09 787.54 -386.54 16.473 1 4.934e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The comparison with the SFO fit without covariate effect confirms that considering the soil pH improves the model, both by comparison of AIC and BIC and by the likelihood ratio test.
plot(sfo_pH)
Endpoints for a model with covariates are by default calculated for the median of the covariate values. This quantile can be adapted, or a specific covariate value can be given as shown below.
endpoints(sfo_pH)$covariates
pH
50% 5.75
$distimes
DT50 DT90
meso 18.52069 61.52441
endpoints(sfo_pH, covariate_quantile = 0.9)$covariates
pH
90% 7.13
$distimes
DT50 DT90
meso 8.237019 27.36278$covariates
pH
User 7
$distimes
DT50 DT90
meso 8.89035 29.5331FOMC
fomc_pH <- saem(f_sep_const["FOMC", ], no_random_effect = "meso_0", covariates = pH,
covariate_models = list(log_alpha ~ pH))| est. | lower | upper | |
|---|---|---|---|
| meso_0 | 92.84 | 90.75 | 94.93 |
| log_alpha | -2.21 | -3.49 | -0.92 |
| beta_pH(log_alpha) | 0.58 | 0.37 | 0.79 |
| log_beta | 4.21 | 3.44 | 4.99 |
| a.1 | 5.03 | 4.32 | 5.73 |
| SD.log_alpha | 0.00 | -23.77 | 23.78 |
| SD.log_beta | 0.37 | 0.01 | 0.74 |
As in the case of SFO, the confidence interval of the slope parameter
(here beta_pH(log_alpha)) quantifying the influence of soil
pH does not include zero, and the model comparison clearly indicates
that the model with covariate influence is preferable. However, the
random effect for alpha is not well-defined any more after
inclusion of the covariate effect (the confidence interval of
SD.log_alpha includes zero).
illparms(fomc_pH)[1] "sd(log_alpha)"Therefore, the model is updated without this random effect, and no ill-defined parameters remain.
anova(f_saem_2[["FOMC", "const"]], fomc_pH, fomc_pH_2, test = TRUE)Data: 116 observations of 1 variable(s) grouped in 18 datasets
npar AIC BIC Lik Chisq Df Pr(>Chisq)
f_saem_2[["FOMC", "const"]] 5 783.25 787.71 -386.63
fomc_pH_2 6 767.49 772.83 -377.75 17.762 1 2.503e-05 ***
fomc_pH 7 770.07 776.30 -378.04 0.000 1 1
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Model comparison indicates that including pH dependence significantly improves the fit, and that the reduced model with covariate influence results in the most preferable FOMC fit.
| est. | lower | upper | |
|---|---|---|---|
| meso_0 | 93.05 | 90.98 | 95.13 |
| log_alpha | -2.91 | -4.18 | -1.63 |
| beta_pH(log_alpha) | 0.66 | 0.44 | 0.87 |
| log_beta | 3.95 | 3.29 | 4.62 |
| a.1 | 4.98 | 4.28 | 5.68 |
| SD.log_beta | 0.40 | 0.26 | 0.54 |
plot(fomc_pH_2)
endpoints(fomc_pH_2)$covariates
pH
50% 5.75
$distimes
DT50 DT90 DT50back
meso 17.30248 82.91343 24.95943$covariates
pH
User 7
$distimes
DT50 DT90 DT50back
meso 6.986239 27.02927 8.136621DFOP
In the DFOP fits without covariate effects, random effects for two
degradation parameters (k2 and g) were
identifiable.
| est. | lower | upper | |
|---|---|---|---|
| meso_0 | 93.61 | 91.58 | 95.63 |
| log_k1 | -1.53 | -2.27 | -0.79 |
| log_k2 | -3.42 | -3.73 | -3.11 |
| g_qlogis | -1.67 | -2.57 | -0.77 |
| a.1 | 4.74 | 4.02 | 5.45 |
| SD.log_k2 | 0.60 | 0.38 | 0.81 |
| SD.g_qlogis | 0.94 | 0.33 | 1.54 |
A fit with pH dependent degradation parameters was obtained by
excluding the same random effects as in the refined DFOP fit without
covariate influence, and including covariate models for the two
identifiable parameters k2 and g.
dfop_pH <- saem(f_sep_const["DFOP", ], no_random_effect = c("meso_0", "log_k1"),
covariates = pH,
covariate_models = list(log_k2 ~ pH, g_qlogis ~ pH))The corresponding parameters for the influence of soil pH are
beta_pH(log_k2) for the influence of soil pH on
k2, and beta_pH(g_qlogis) for its influence on
g.
| est. | lower | upper | |
|---|---|---|---|
| meso_0 | 92.84 | 90.85 | 94.84 |
| log_k1 | -2.82 | -3.09 | -2.54 |
| log_k2 | -11.48 | -15.32 | -7.64 |
| beta_pH(log_k2) | 1.31 | 0.69 | 1.92 |
| g_qlogis | 3.13 | 0.47 | 5.80 |
| beta_pH(g_qlogis) | -0.57 | -1.04 | -0.09 |
| a.1 | 4.96 | 4.26 | 5.65 |
| SD.log_k2 | 0.76 | 0.47 | 1.05 |
| SD.g_qlogis | 0.01 | -9.96 | 9.97 |
illparms(dfop_pH)[1] "sd(g_qlogis)"Confidence intervals for neither of them include zero, indicating a
significant difference from zero. However, the random effect for
g is now ill-defined. The fit is updated without this
ill-defined random effect.
dfop_pH_2 <- update(dfop_pH,
no_random_effect = c("meso_0", "log_k1", "g_qlogis"))
illparms(dfop_pH_2)[1] "beta_pH(g_qlogis)"Now, the slope parameter for the pH effect on g is
ill-defined. Therefore, another attempt is made without the
corresponding covariate model.
dfop_pH_3 <- saem(f_sep_const["DFOP", ], no_random_effect = c("meso_0", "log_k1"),
covariates = pH,
covariate_models = list(log_k2 ~ pH))
illparms(dfop_pH_3)[1] "sd(g_qlogis)"As the random effect for g is again ill-defined, the fit
is repeated without it.
dfop_pH_4 <- update(dfop_pH_3, no_random_effect = c("meso_0", "log_k1", "g_qlogis"))
illparms(dfop_pH_4)While no ill-defined parameters remain, model comparison suggests
that the previous model dfop_pH_2 with two pH dependent
parameters is preferable, based on information criteria as well as based
on the likelihood ratio test.
anova(f_saem_2[["DFOP", "const"]], dfop_pH, dfop_pH_2, dfop_pH_3, dfop_pH_4)Data: 116 observations of 1 variable(s) grouped in 18 datasets
npar AIC BIC Lik
f_saem_2[["DFOP", "const"]] 7 782.94 789.18 -384.47
dfop_pH_4 7 767.35 773.58 -376.68
dfop_pH_2 8 765.14 772.26 -374.57
dfop_pH_3 8 769.00 776.12 -376.50
dfop_pH 9 769.10 777.11 -375.55
anova(dfop_pH_2, dfop_pH_4, test = TRUE)Data: 116 observations of 1 variable(s) grouped in 18 datasets
npar AIC BIC Lik Chisq Df Pr(>Chisq)
dfop_pH_4 7 767.35 773.58 -376.68
dfop_pH_2 8 765.14 772.26 -374.57 4.2153 1 0.04006 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1When focussing on parameter identifiability using the test if the
confidence interval includes zero, dfop_pH_4 would still be
the preferred model. However, it should be kept in mind that parameter
confidence intervals are constructed using a simple linearisation of the
likelihood. As the confidence interval of the random effect for
g only marginally includes zero, it is suggested that this
is acceptable, and that dfop_pH_2 can be considered the
most preferable model.
plot(dfop_pH_2)
endpoints(dfop_pH_2)$covariates
pH
50% 5.75
$distimes
DT50 DT90 DT50back DT50_k1 DT50_k2
meso 18.36876 73.51841 22.13125 4.191901 23.98672$covariates
pH
User 7
$distimes
DT50 DT90 DT50back DT50_k1 DT50_k2
meso 8.346428 28.34437 8.532507 4.191901 8.753618SFORB
sforb_pH <- saem(f_sep_const["SFORB", ], no_random_effect = c("meso_free_0", "log_k_meso_free_bound"),
covariates = pH,
covariate_models = list(log_k_meso_free ~ pH, log_k_meso_bound_free ~ pH))| est. | lower | upper | |
|---|---|---|---|
| meso_free_0 | 93.42 | 91.32 | 95.52 |
| log_k_meso_free | -5.37 | -6.94 | -3.81 |
| beta_pH(log_k_meso_free) | 0.42 | 0.18 | 0.67 |
| log_k_meso_free_bound | -3.49 | -4.92 | -2.05 |
| log_k_meso_bound_free | -9.98 | -19.22 | -0.74 |
| beta_pH(log_k_meso_bound_free) | 1.23 | -0.21 | 2.67 |
| a.1 | 4.90 | 4.18 | 5.63 |
| SD.log_k_meso_free | 0.35 | 0.23 | 0.47 |
| SD.log_k_meso_bound_free | 0.13 | -1.95 | 2.20 |
The confidence interval of
beta_pH(log_k_meso_bound_free) includes zero, indicating
that the influence of soil pH on k_meso_bound_free cannot
reliably be quantified. Also, the confidence interval for the random
effect on this parameter (SD.log_k_meso_bound_free)
includes zero.
Using the illparms function, these ill-defined
parameters can be found more conveniently.
illparms(sforb_pH)[1] "sd(log_k_meso_bound_free)" "beta_pH(log_k_meso_bound_free)"To remove the ill-defined parameters, a second variant of the SFORB model with pH influence is fitted. No ill-defined parameters remain.
sforb_pH_2 <- update(sforb_pH,
no_random_effect = c("meso_free_0", "log_k_meso_free_bound", "log_k_meso_bound_free"),
covariate_models = list(log_k_meso_free ~ pH))
illparms(sforb_pH_2)The model comparison of the SFORB fits includes the refined model without covariate effect, and both versions of the SFORB fit with covariate effect.
anova(f_saem_2[["SFORB", "const"]], sforb_pH, sforb_pH_2, test = TRUE)Data: 116 observations of 1 variable(s) grouped in 18 datasets
npar AIC BIC Lik Chisq Df Pr(>Chisq)
f_saem_2[["SFORB", "const"]] 7 783.40 789.63 -384.70
sforb_pH_2 7 770.94 777.17 -378.47 12.4616 0
sforb_pH 9 768.81 776.83 -375.41 6.1258 2 0.04675 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The first model including pH influence is preferable based on information criteria and the likelihood ratio test. However, as it is not fully identifiable, the second model is selected.
| est. | lower | upper | |
|---|---|---|---|
| meso_free_0 | 93.32 | 91.16 | 95.48 |
| log_k_meso_free | -6.15 | -7.43 | -4.86 |
| beta_pH(log_k_meso_free) | 0.54 | 0.33 | 0.75 |
| log_k_meso_free_bound | -3.80 | -5.20 | -2.40 |
| log_k_meso_bound_free | -2.95 | -4.26 | -1.64 |
| a.1 | 5.08 | 4.38 | 5.79 |
| SD.log_k_meso_free | 0.33 | 0.22 | 0.45 |
plot(sforb_pH_2)
endpoints(sforb_pH_2)$covariates
pH
50% 5.75
$ff
meso_free
1
$SFORB
meso_b1 meso_b2 meso_g
0.09735824 0.02631699 0.31602120
$distimes
DT50 DT90 DT50back DT50_meso_b1 DT50_meso_b2
meso 16.86549 73.15824 22.02282 7.119554 26.33839$covariates
pH
User 7
$ff
meso_free
1
$SFORB
meso_b1 meso_b2 meso_g
0.13315233 0.03795988 0.61186191
$distimes
DT50 DT90 DT50back DT50_meso_b1 DT50_meso_b2
meso 7.932495 36.93311 11.11797 5.205671 18.26HS
hs_pH <- saem(f_sep_const["HS", ], no_random_effect = c("meso_0"),
covariates = pH,
covariate_models = list(log_k1 ~ pH, log_k2 ~ pH, log_tb ~ pH))| est. | lower | upper | |
|---|---|---|---|
| meso_0 | 93.33 | 91.47 | 95.19 |
| log_k1 | -5.81 | -7.27 | -4.36 |
| beta_pH(log_k1) | 0.47 | 0.23 | 0.72 |
| log_k2 | -6.80 | -8.76 | -4.83 |
| beta_pH(log_k2) | 0.54 | 0.21 | 0.87 |
| log_tb | 3.25 | 1.25 | 5.25 |
| beta_pH(log_tb) | -0.10 | -0.43 | 0.23 |
| a.1 | 4.49 | 3.78 | 5.21 |
| SD.log_k1 | 0.37 | 0.24 | 0.51 |
| SD.log_k2 | 0.29 | 0.10 | 0.48 |
| SD.log_tb | 0.25 | -0.07 | 0.57 |
illparms(hs_pH)[1] "sd(log_tb)" "beta_pH(log_tb)"According to the output of the illparms function, the
random effect on the break time tb cannot reliably be
quantified, neither can the influence of soil pH on tb. The
fit is repeated without the corresponding covariate model, and no
ill-defined parameters remain.
Model comparison confirms that this model is preferable to the fit without covariate influence, and also to the first version with covariate influence.
anova(f_saem_2[["HS", "const"]], hs_pH, hs_pH_2, test = TRUE)Data: 116 observations of 1 variable(s) grouped in 18 datasets
npar AIC BIC Lik Chisq Df Pr(>Chisq)
f_saem_2[["HS", "const"]] 8 780.08 787.20 -382.04
hs_pH_2 10 766.47 775.37 -373.23 17.606 2 0.0001503 ***
hs_pH 11 769.80 779.59 -373.90 0.000 1 1.0000000
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1| est. | lower | upper | |
|---|---|---|---|
| meso_0 | 93.33 | 91.50 | 95.15 |
| log_k1 | -5.68 | -7.09 | -4.27 |
| beta_pH(log_k1) | 0.46 | 0.22 | 0.69 |
| log_k2 | -6.61 | -8.34 | -4.88 |
| beta_pH(log_k2) | 0.50 | 0.21 | 0.79 |
| log_tb | 2.70 | 2.33 | 3.08 |
| a.1 | 4.45 | 3.74 | 5.16 |
| SD.log_k1 | 0.36 | 0.22 | 0.49 |
| SD.log_k2 | 0.23 | 0.02 | 0.43 |
| SD.log_tb | 0.55 | 0.25 | 0.85 |
plot(hs_pH_2)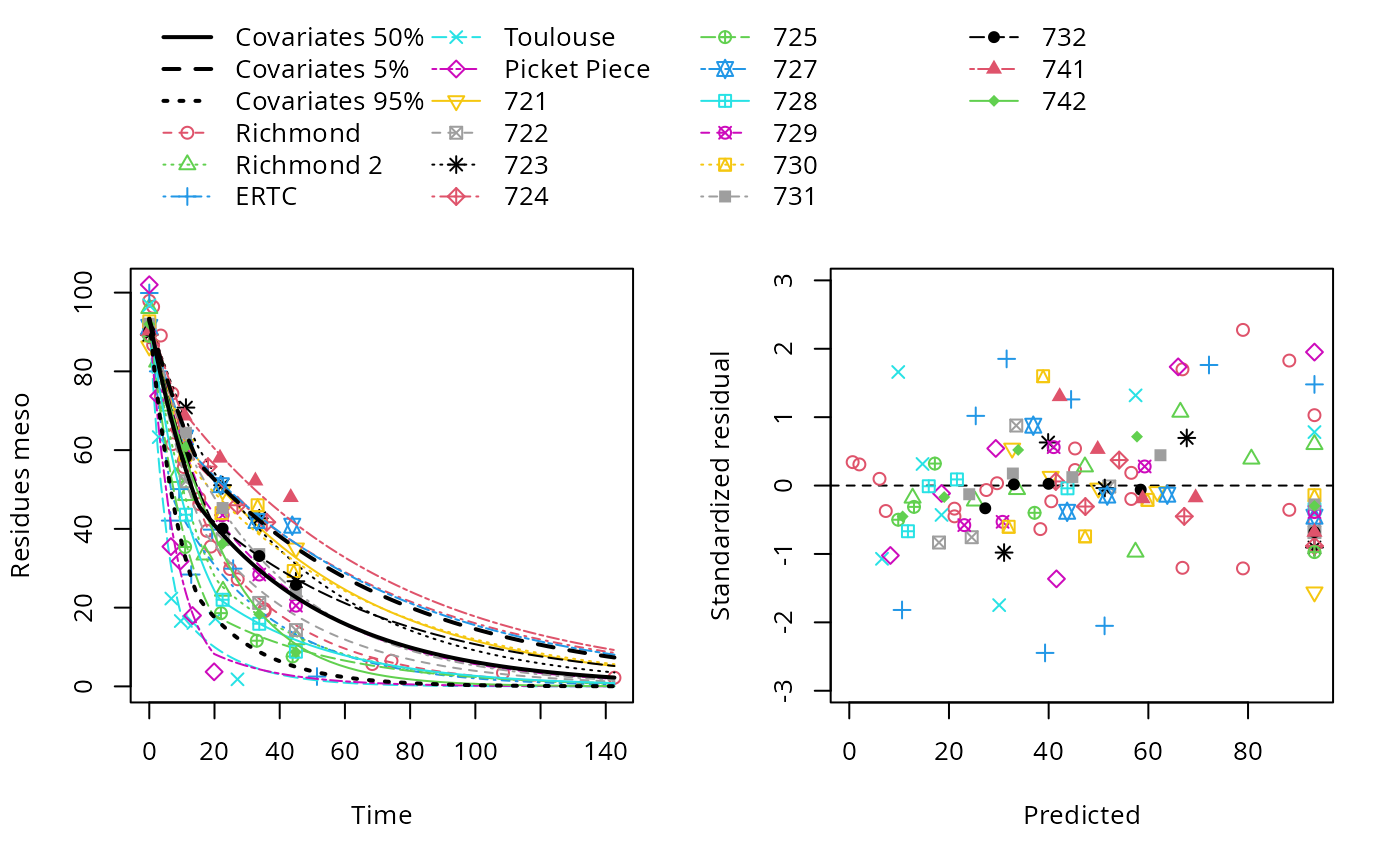
endpoints(hs_pH_2)$covariates
pH
50% 5.75
$distimes
DT50 DT90 DT50back DT50_k1 DT50_k2
meso 14.68725 82.45287 24.82079 14.68725 29.29299$covariates
pH
User 7
$distimes
DT50 DT90 DT50back DT50_k1 DT50_k2
meso 8.298536 38.85371 11.69613 8.298536 15.71561Comparison across parent models
After model reduction for all models with pH influence, they are compared with each other.
anova(sfo_pH, fomc_pH_2, dfop_pH_2, dfop_pH_4, sforb_pH_2, hs_pH_2)Data: 116 observations of 1 variable(s) grouped in 18 datasets
npar AIC BIC Lik
sfo_pH 5 783.09 787.54 -386.54
fomc_pH_2 6 767.49 772.83 -377.75
dfop_pH_4 7 767.35 773.58 -376.68
sforb_pH_2 7 770.94 777.17 -378.47
dfop_pH_2 8 765.14 772.26 -374.57
hs_pH_2 10 766.47 775.37 -373.23The DFOP model with pH influence on k2 and
g and a random effect only on k2 is finally
selected as the best fit.
The endpoints resulting from this model are listed below. Please refer to the Appendix for a detailed listing.
endpoints(dfop_pH_2)$covariates
pH
50% 5.75
$distimes
DT50 DT90 DT50back DT50_k1 DT50_k2
meso 18.36876 73.51841 22.13125 4.191901 23.98672$covariates
pH
User 7
$distimes
DT50 DT90 DT50back DT50_k1 DT50_k2
meso 8.346428 28.34437 8.532507 4.191901 8.753618Conclusions
These evaluations demonstrate that covariate effects can be included for all types of parent degradation models. These models can then be further refined to make them fully identifiable.
Appendix
Session info
R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Debian GNU/Linux 12 (bookworm)
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.11.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.11.0
locale:
[1] LC_CTYPE=de_DE.UTF-8 LC_NUMERIC=C
[3] LC_TIME=de_DE.UTF-8 LC_COLLATE=de_DE.UTF-8
[5] LC_MONETARY=de_DE.UTF-8 LC_MESSAGES=de_DE.UTF-8
[7] LC_PAPER=de_DE.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=de_DE.UTF-8 LC_IDENTIFICATION=C
time zone: Europe/Berlin
tzcode source: system (glibc)
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] saemix_3.3 npde_3.5 knitr_1.49 mkin_1.2.10
loaded via a namespace (and not attached):
[1] gtable_0.3.6 jsonlite_1.8.9 dplyr_1.1.4 compiler_4.4.2
[5] tidyselect_1.2.1 gridExtra_2.3 jquerylib_0.1.4 systemfonts_1.1.0
[9] scales_1.3.0 textshaping_0.4.1 yaml_2.3.10 fastmap_1.2.0
[13] lattice_0.22-6 ggplot2_3.5.1 R6_2.5.1 generics_0.1.3
[17] lmtest_0.9-40 MASS_7.3-61 htmlwidgets_1.6.4 tibble_3.2.1
[21] desc_1.4.3 munsell_0.5.1 bslib_0.8.0 pillar_1.9.0
[25] rlang_1.1.4 utf8_1.2.4 cachem_1.1.0 xfun_0.49
[29] fs_1.6.5 sass_0.4.9 cli_3.6.3 pkgdown_2.1.1
[33] magrittr_2.0.3 digest_0.6.37 grid_4.4.2 mclust_6.1.1
[37] lifecycle_1.0.4 nlme_3.1-166 vctrs_0.6.5 evaluate_1.0.1
[41] glue_1.8.0 ragg_1.3.3 zoo_1.8-12 fansi_1.0.6
[45] colorspace_2.1-1 rmarkdown_2.29 pkgconfig_2.0.3 tools_4.4.2
[49] htmltools_0.5.8.1